Description
Overview

Flowers are very old friends to humans. By stimulating our sense of sight and smell, flowers have a great
impact on our well-being. Flowers add color to various scenes of our daily lives as gifts and decorations
and are also used in a special role in milestone events of life such as weddings and funerals.
If we think about the bigger picture, flowers are even more deeply tied to our lives. Our lives totally depend on flowers, because food comes from flowering plants.
If we think about the bigger picture, flowers are even more deeply tied to our lives. Our lives totally depend on flowers, because food comes from flowering plants.
However, enormous amounts of such valuable flowers are lost before they reach us as products.
Plant diseases cause serious problems during cultivation. Plants produce 80% of food, however, 20 to
40% of
them are lost by plant pests, according to the FAO’s report[1].

Today, virus and viroid infections are spreading in plant production, causing growth failure,
dwarfing, and blemishes in plant pigmentation, which reduces the efficiency of plant production. Plant
diseases cost the global economy around US$220 billion per year[2].

For example, when dahlia is infected by a virus (DMV: Dahlia mosaic
virus), leaf mosaic symptoms and systemic chlorosis are observed, and growth of the entire plant is
suppressed [3].
When tomatoes are infected with a virus (TSWV: Tomato spotted wilt virus), the fruits show deformities characteristically, and the surface also shows uneven ripening and bulges [4].
Likewise, if cucumbers are infected with the virus (ZYMV: Zucchini yellow mosaic virus), they will become stunted and the fruit will have knobby areas which cause embossed deformation and irregular skin coloring [5]. Such physical defects result in vegetables that are rejected by consumers, resulting in unnecessary waste.
When tomatoes are infected with a virus (TSWV: Tomato spotted wilt virus), the fruits show deformities characteristically, and the surface also shows uneven ripening and bulges [4].
Likewise, if cucumbers are infected with the virus (ZYMV: Zucchini yellow mosaic virus), they will become stunted and the fruit will have knobby areas which cause embossed deformation and irregular skin coloring [5]. Such physical defects result in vegetables that are rejected by consumers, resulting in unnecessary waste.
There is no cure for these viral and viroid infections, and the only current countermeasure is to find
diseased plants in the early stages of infection and remove them. However, it is difficult to visually
select healthy plants because of the varying degrees of disease symptoms and the long periods of latent
infection or presentation of disease symptoms[6].
This year iGEM Kyoto aimed to tackle this environmental challenge using synthetic biology.
Based on the advice of experts, we built our system to target the dahlia virus. Our goal is to establish an effective and practical solution which may be used by farmers to combat dahlia virus infection. Moreover, we developed an efficient biomolecule producing system called “BLOOM”, and also proved it conceptually by modeling and simulations. This will support not only our solution for plant disease but also any production of biomolecules.
This year iGEM Kyoto aimed to tackle this environmental challenge using synthetic biology.
Based on the advice of experts, we built our system to target the dahlia virus. Our goal is to establish an effective and practical solution which may be used by farmers to combat dahlia virus infection. Moreover, we developed an efficient biomolecule producing system called “BLOOM”, and also proved it conceptually by modeling and simulations. This will support not only our solution for plant disease but also any production of biomolecules.
1. Solutions for Flower Problems
Infectious Disease Detection
How can we find an infected flowering plant in a vast farm? Can drones analyze the whole field by scanning
from height and let us know where diseased plants are? Unfortunately, no such technologies are currently
available. Even image diagnosis by machine learning is still in the rudimentary stages due to the lack of a
sufficient database of images of diseased leaves[7][8][9].
The current method of virus testing is to collect bulbs before they are planted by using qPCR (or qRT-PCR) for nucleic acid amplification and detection at agricultural experiment stations[10]. Such quality controls of bulbs eliminate the virus from newly planted ones. However, growing plants can be infected by a variety of wild viruses transmitted by insects such as thrips. Such natural infections are hardly detected by the human eyes, causing spread of the viruses in the field[11].
The current method of virus testing is to collect bulbs before they are planted by using qPCR (or qRT-PCR) for nucleic acid amplification and detection at agricultural experiment stations[10]. Such quality controls of bulbs eliminate the virus from newly planted ones. However, growing plants can be infected by a variety of wild viruses transmitted by insects such as thrips. Such natural infections are hardly detected by the human eyes, causing spread of the viruses in the field[11].
To solve this problem, we developed a multi-layered system that enables farmers to diagnose
viral infection
on-site. This is a system to detect diseased plants more easily and accurately than ever before by
using
machine learning for image diagnosis combined with CRISPR-Cas12a assisted (RT)-LAMP for
molecular
validation.


As an image diagnosis tool, we developed a software called "DLAEMON (Diseased Leaves
Assessment by Efficient
Machine-learning On Neural network)" to discern infected leaves and healthy leaves by machine learning.

The samples are easily tested by CRISPR-Cas12a assisted (RT)-LAMP, which does not
require a thermal cycler. To allow farmers to check the results on-site, we created a hardware
“DLAMI (DLA's Machine Interface)”.
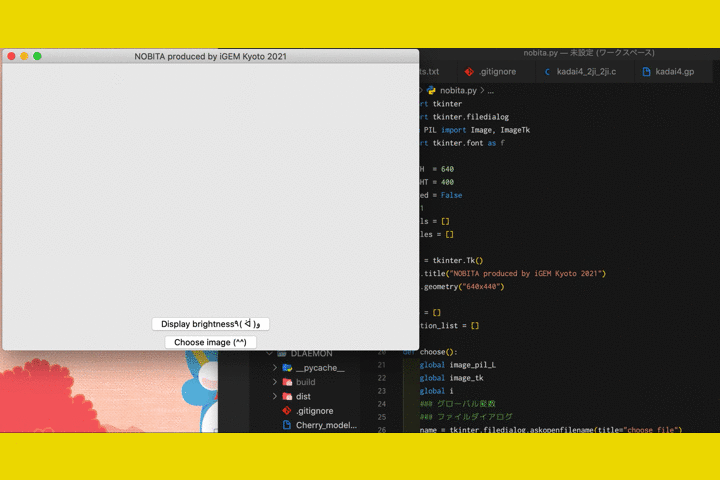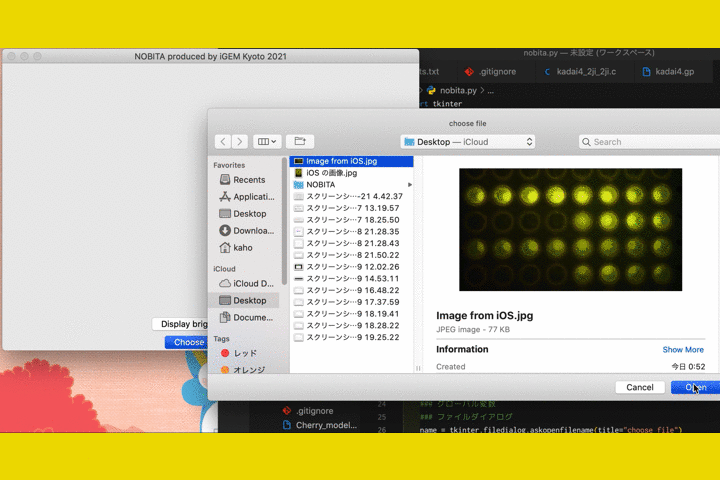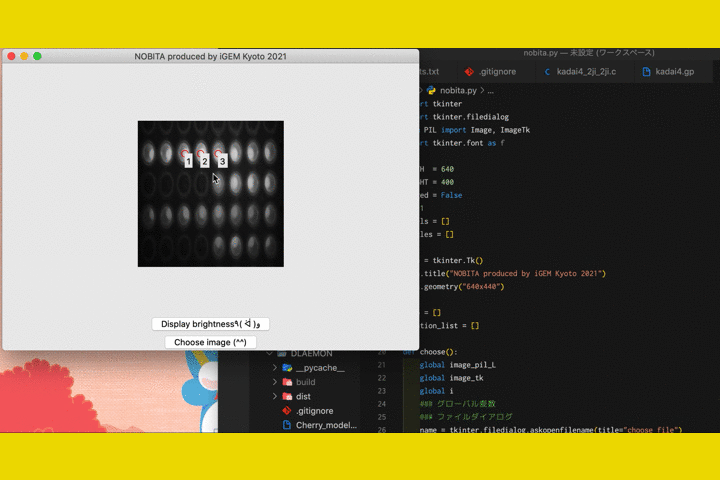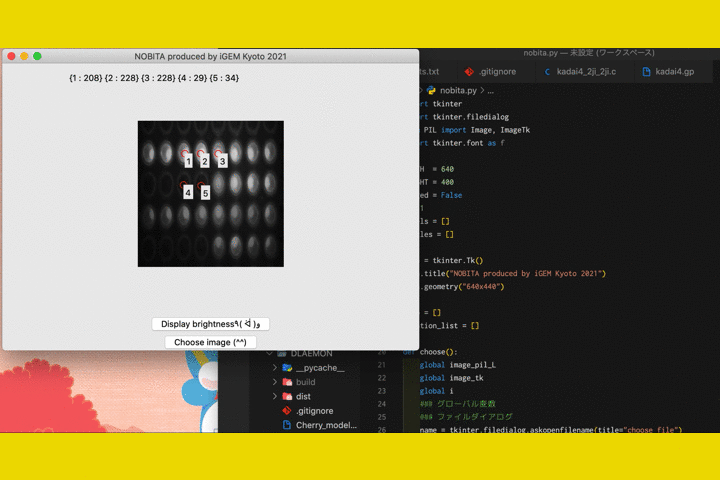
We
also developed a software called "NOBITA (Numericalization Of Brightness of Image for one-Touch
Assessment)"
to read out the fluorescence intensity.

We expect to achieve the “DLA cycle”: a feed-forward loop drives DLAEMON’s
evolution by linking a
combination of images used by DLAEMON and the fluorescence intensity determined by NOBITA. A newly
acquired dataset will be used for the improvement of DLAEMON, which will predict infected leaves more
precisely and increase the efficiency of diseased leaves detection. Repeating this cycle, DLAEMON will
become precise enough. One day, a fully evolved version of DLAEMON will not need enzyme reactions for the
test any more. This will allow us more flexible applications, for example, drones with DLAEMON can scan
whole fields by taking images then let farmers know areas to be diagnosed by DLAMI.
Multifaceted support for flower production
Thrips are known as a vehiculum of plant virus, as they spread infections. In addition to the improvement of
virus detection, we also dealt with the causes of virus spread. Frankliniella occidentalis: one species of
thrips, is causing serious damage to farmers because it spreads Tomato spotted wilt virus (TSWV), besides,
it has multiple drug resistance. We came up with the idea of using RNAi to kill this species in
fields[12].

Frankliniella occidentalis is a mediator of TSWV

An idea of RNAi
Moreover, we also took on the challenge of reducing the environmental impact of STS (silver thiosulfate),
which is currently used to stop the aging of flowers by ethylene[13], by replacing it with the biopeptide
NOP-1.
Finally, to extend the life of cut flowers, we attempted to delay the blockage of the vessels that cause wilting of cut flowers by administering antimicrobial peptides and biofilm-degrading enzymes that prevent the growth of bacteria[14][15],.
Finally, to extend the life of cut flowers, we attempted to delay the blockage of the vessels that cause wilting of cut flowers by administering antimicrobial peptides and biofilm-degrading enzymes that prevent the growth of bacteria[14][15],.
Our goal
- Detection of a diseased plant by image diagnosis
- Enzymatic reaction test using (RT)-LAMP in combination with Cas12a
- Development of hardware for measuring fluorescence
- Development of software to quantify fluorescence
- Production of various biomolecules involved in flower production and distribution
Thus, our project aims to solve the problems of the flower industry by using microorganisms to produce a variety of biomaterials. For this project to be an environmentally sustainable activity, the production system for these materials itself must be efficient and have a low environmental load. So we reassessed existing biomaterial producing systems and developed a novel type of continuous culture system which enables an efficient synthesis of target molecules.

Biomolecular Production Platform
Producing biomolecules efficiently is one of the biggest challenges for synthetic biology and
sustainability.
The methods actually used in bioprocessing today can be broadly divided into two categories: batch cultures, in which cells are collected after each expression cycle, and continuous cultures, in which bacteria are collected continuously while synthesizing products. In the latter, high-density culture can produce more efficient products per unit volume. Although, in continuous culture, complex regulation such as multiple stepwise control of protein expression is unavailable.
The methods actually used in bioprocessing today can be broadly divided into two categories: batch cultures, in which cells are collected after each expression cycle, and continuous cultures, in which bacteria are collected continuously while synthesizing products. In the latter, high-density culture can produce more efficient products per unit volume. Although, in continuous culture, complex regulation such as multiple stepwise control of protein expression is unavailable.
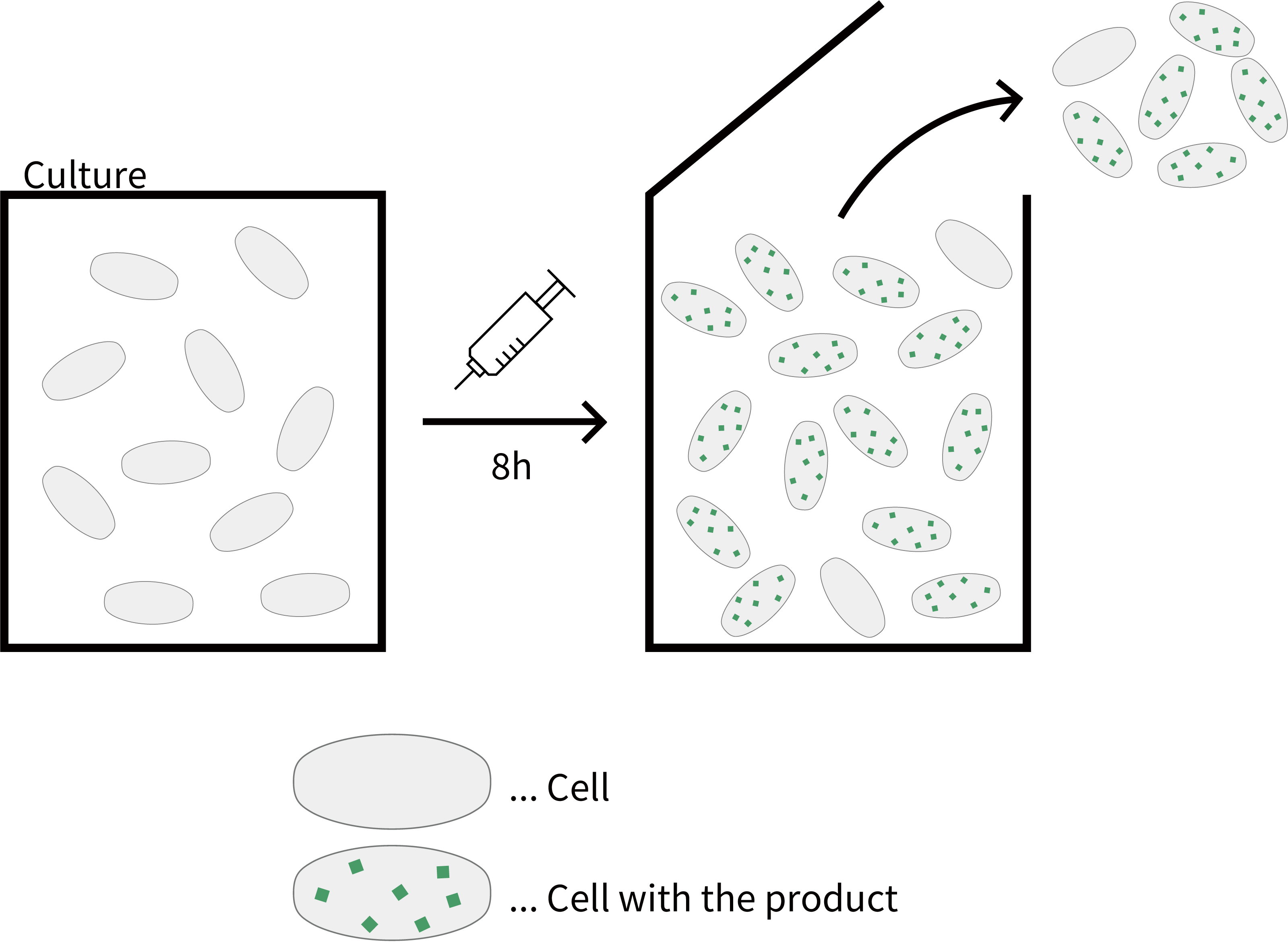
Fig. Batch culture

Fig. Continuous culture
Our system “BLOOM”, inspired by a cell
differentiation system, overcame this weak point and allows for multi-level complex control.
Efficient and stable production platform
using an asymmetric plasmid partitioning system
using an asymmetric plasmid partitioning system


Fig. Comparison between batch culture and continuous culture
Division of labor offers efficiency for production. For example, in multicellular organisms, cell
differentiation leads to a "division of labor". Populations of differentiated cells are capable of
producing a wide variety of molecules beyond the levels achievable with populations of a single type of
cell.
Therefore, we decided to revise the continuous culture system by utilizing a bacterial differentiation system based on “asymmetric plasmid partitioning” or APP. The APP system was reported in 2019[16]. Usually, plasmids are divided into daughter cells equally after cell division. In APP, one of at least two different types of plasmids are inherited into only one of the daughter cells. Cells with a full complement of plasmids are considered “stem cells”. Cells which retain only a partial complement of plasmids become “differentiated cells”.
Therefore, we decided to revise the continuous culture system by utilizing a bacterial differentiation system based on “asymmetric plasmid partitioning” or APP. The APP system was reported in 2019[16]. Usually, plasmids are divided into daughter cells equally after cell division. In APP, one of at least two different types of plasmids are inherited into only one of the daughter cells. Cells with a full complement of plasmids are considered “stem cells”. Cells which retain only a partial complement of plasmids become “differentiated cells”.

Differenciation of cells in APP system
In our system, only differentiated cells produce target biomolecules. Stem cells
spontaneously produce
differentiated cells, and maintain the population during continuous culture. We introduced two kinds of
repressors each with a different degradation rate so that differentiated cells achieve stepwise
expression
of several genes. Also, we built a mathematical model of this system and investigated the
requirements for
operation.

Among several parameters explored, our simulation results showed manipulation of degradation rates of
repressors is critical to control a stepwise expression of multiple genes. Based on this result, we made a
collection of SsrA mutants. SsrA is a degradation-inducing tag in E. coli. We introduced various mutations
into the C-terminus of this tag to gain mutant SsrA with different degradation rates (See the Parts page)[17][18]. This
collection not only expands usage of our system but also helps any implementation of protein expression in
E. coli which needs fine-tuning of degradation rate.
What could be a possible application if we can program the timing for the expression of multiple genes with BLOOM? As an example of applications, we simulated E. coli culture programmed to:
In this system, GFP expressing cells will be aggregated and easily collected from a continuous culture by filtration. In this system, by using cell-autonomous control, we can collect cells right after they complete GFP production. This is a clear advantage of BLOOM from ordinary continuous culture, because normal continuous culture randomly collects cells containing less amount of GFP or GFP produced a long time before the harvest, resulting in the potential quality problem of the harvested protein, and lot-to-lot variations.

Furthermore, with BLOOM, we can program a system to induce E. coli to self-destruct after the expression and harvest steps. Such a program will be useful for implementing a system with guaranteed biosafety. Please refer to the links for more information on the various applications and the underlying technologies that support them.
What could be a possible application if we can program the timing for the expression of multiple genes with BLOOM? As an example of applications, we simulated E. coli culture programmed to:
- produce amolecule of target (GFP),
- express a protein which induces cell-cell aggregation for harvest[19].
In this system, GFP expressing cells will be aggregated and easily collected from a continuous culture by filtration. In this system, by using cell-autonomous control, we can collect cells right after they complete GFP production. This is a clear advantage of BLOOM from ordinary continuous culture, because normal continuous culture randomly collects cells containing less amount of GFP or GFP produced a long time before the harvest, resulting in the potential quality problem of the harvested protein, and lot-to-lot variations.

A possible application of BLOOM
Furthermore, with BLOOM, we can program a system to induce E. coli to self-destruct after the expression and harvest steps. Such a program will be useful for implementing a system with guaranteed biosafety. Please refer to the links for more information on the various applications and the underlying technologies that support them.
References
- Food and Agriculture Organization of the United Nations, The future of food and agriculture Trends and Challenges, https://www.fao.org/3/i6583e/I6583E.pdf
- Research Papers (2021). Pathogens, precipitation and produce prices. Nat. Clim. Chang. 11, 635–635.
- Pappu, H.R., Wyatt, S.D., and Druffel, K.L. (2005) "Dahlia mosaic virus: Molecular detection and distribution in Dahlia in the United States", HortScience 40, 697–699.
- Sether, D.M., DeAngelis, J.D., and Oregon State University. Agricultural Experiment Station (1992) Tomato spotted wilt virus host list and bibliography.
- Desbiez, C., and Lecoq, H. (1997) "Zucchini yellow mosaic virus", Plant Pathol. 46, 809–829.
- Rubio, L., Galipienso, L., and Ferriol, I. (2020). Detection of Plant Viruses and Disease Management: Relevance of Genetic Diversity and Evolution. Front. Plant Sci. 11, 1092.
- Mohanty, S.P., Hughes, D.P., and Salathé, M. (2016). Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 7, 1419.
- Liu, J., and Wang, X. (2021). Plant diseases and pests detection based on deep learning: a review. Plant Methods 17, 22.
- Khan, R.U., Khan, K., Albattah, W., and Qamar, A.M. (2021). Image-Based Detection of Plant Diseases: From Classical Machine Learning to Deep Learning Journey. Proc. Int. Wirel. Commun. Mob. Comput. Conf. 2021.
- [JAPANESE] 球根増殖コンソーシアム ダリアのウイルス・ウイロイド病 診断マニュアル (2019)
- Nilon, A., Robinson, K., Pappu, H.R., and Mitter, N. (2021). Current Status and Potential of RNA Interference for the Management of Tomato Spotted Wilt Virus and Thrips Vectors. Pathogens 10.
- Guan, R., Chu, D., Han, X., Miao, X., and Li, H. (2021). Advances in the Development of Microbial Double-Stranded RNA Production Systems for Application of RNA Interference in Agricultural Pest Control. Front Bioeng Biotechnol 9, 753790.
- Hoppen, C., Müller, L., Albrecht, A.C., and Groth, G. (2019). The NOP-1 peptide derived from the central regulator of ethylene signaling EIN2 delays floral senescence in cut flowers. Sci. Rep. 9, 1287.
- Mohammadi, M., Aelaei, M., and Saidi, M. (2020). Antibacterial properties of Scrophularia striata Boiss. (Tashenehdari) extract on vase life improvement in “Stanza” and “Pink Elegance” gerbera cut flowers. Biocatal. Agric. Biotechnol. 28, 101738.
- Put, H.M.C. (1990). Micro-organisms from freshly harvested cut flower stems and developing during the vase life of chrysanthemum, gerbera and rose cultivars. Sci. Hortic. 43, 129–144.
- Molinari, S., Shis, D.L., Bhakta, S.P., Chappell, J., Igoshin, O.A., and Bennett, M.R. (2019) "A synthetic system for asymmetric cell division in Escherichia coli", Nat. Chem. Biol. 15, 917–924.
- Karzai, A.W., Roche, E.D., and Sauer, R.T. (2000). The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7, 449–455.
- Flynn, J.M., Levchenko, I., Seidel, M., Wickner, S.H., Sauer, R.T., and Baker, T.A. (2001). Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. U. S. A. 98, 10584–10589.
- Ageorges, V., Schiavone, M., Jubelin, G., Caccia, N., Ruiz, P., Chafsey, I., Bailly, X., Dague, E., Leroy, S., Paxman, J., et al. (2019). Differential homotypic and heterotypic interactions of antigen 43 (Ag43) variants in autotransporter-mediated bacterial autoaggregation. Sci. Rep. 9, 11100.
