Proof Of Concept
- Click to visit -
Overview
Toward the implementation of our project in a relevant context, we completed proof
of
concept experiments. First, to solve the problem of viral infection in flowering plants, our
experiments
and prototypes include an enzymatic detection method, image-based software AI, and an integrated
hardware
system. Additionally, we performed experiments to address the control of plant pests, as well as
conceptualized and modeled an improved biomolecule production system with broad applications in
our own
project and general bioprocesses. The following results establish proof of concept for each
component and
the project as a whole (see also
Implementation).


Fig.1 Whole story of Implementation
DLA
1. DLAEMON;
Diseased Leaves Assessment by Efficient Machine-learning On Neural network
Diseased Leaves Assessment by Efficient Machine-learning On Neural network
- Machine learning accurately diagnoses viral infection of
leaves
We have developed DLAEMON, a system for the image diagnosis of infected leaves, as described in
the
Software page. Devising a machine learning method based on neural networks has allowed us to
build
software that can diagnose very efficiently and accurately. The method used is called
fine-tuning, a
technology that enables efficient learning with a limited amount of data by using a neural
network (in our
case, Resnet18) that has already been trained and can be applied to a wide range of data. It is
expected
that further fine-tuning of the AI we developed will make it easier to develop diagnostic
imaging AI for
other types of leaves (see also Software).




Fig.2 Accuracy
The learning curve is as follows: the final accuracy for the training set is 99.9%, the final accuracy for the val set is 99.7%, and the accuracy for the test set is 99.8%.
The learning curve is as follows: the final accuracy for the training set is 99.9%, the final accuracy for the val set is 99.7%, and the accuracy for the test set is 99.8%.

Fig.3 The screen of the application for diagnosis.
This software, DLAEMON, diagnoses images of leaves taken with a smartphone and determines whether they are infected or not. Its user interface looks like this.
This software, DLAEMON, diagnoses images of leaves taken with a smartphone and determines whether they are infected or not. Its user interface looks like this.

Fig.4 Fine-tuning
One of the main features of DLAEMON is the possibility of the extension of this software via
fine-tuning.
This property makes it possible to use DLAEMON not only for the diagnosis of the images we dealt
with here
but also for the detection of viral infections on various plants.
These results show that we can predict viral infections from plant leaf images at an accurate
level, which
enhances the prior probability for the enzymatic virus detection tests presented in the
following pages.
2. Enzymatic Reaction
- RT-LAMP coupled with CRISPR-Cas12a enables quick and sensitive
detection of viral
nucleic acids
Nucleic acid amplification tests using a combination of RT-LAMP and CRISPR-Cas12a have already
been
reported in several papers. These features include the fact that no complicated equipment is
required due
to the isothermal reaction, and that high sensitivity can be obtained. We first verified that
this system
can detect the nucleic acids of plant viruses and proved the concept (see the Engineering page).

Fig.5 A photo showing Fluorescence
The detection of plant viruses was demonstrated to be practicable.
Enzyme reaction was tested with two different guide RNAs. +virus DNA shows more fluorescence compare to negative control.
The detection of plant viruses was demonstrated to be practicable.
Enzyme reaction was tested with two different guide RNAs. +virus DNA shows more fluorescence compare to negative control.
The system we are aiming for is a package of inexpensive and efficient ways of viral detection.
With the
expression and purification of enzymes utilizing BLOOM System, discussed later, we are planning
to supply
the components necessary for tests in the future. We purified the necessary enzymes, Bst DNA
polymerase
and Reversetranscriptase, as recombinants from the parts we prepared by ourselves, and examined
whether
the reaction could be reconstructed. We obtained results with in-house produced enzymes that are
comparable to commercial sources.

Fig.6 Ladder of LAMP reaction with self-made Bst DNA polymerase

Fig.7 The success of reverse transcription with self-made RT
The enzymes for which these functions were demonstrated were also registered as parts, and the
purification methods and reaction conditions were shown in detail on the Results page.
3. DLAMI;
DLA's Machine-Interface
DLA's Machine-Interface
- Our well-designed device is easy enough for farmers to use
We have succeeded in detecting nucleic acids and generating fluorescence, but we need a small
darkroom for
detection before it can be practical in the field. We named this DLAMI and demonstrated that it
can be
made with a 3D printer and placed in the field to detect fluorescence.

Fig.8 DLAMI
Put the tube of the sample in a designated place and close the lid. Turn on the LED and capture the excitation light of the sample through the camera hole.
Put the tube of the sample in a designated place and close the lid. Turn on the LED and capture the excitation light of the sample through the camera hole.
Assuming that it would be used with convenience by farmers, several ingenious measures were
taken during
the designing process. For example, we assumed that it would be photographed with a smartphone,
which is a
common device found in nearly everyone’s pockets. The size of the camera hole and the distance
from the
camera to the sample were designed for this purpose. Photos taken (Fig.5) with this device allow
us to observe
fluorescence to some extent even with the naked eye (see the Hardware page).
4. NOBITA;
Numericalization Of Brightness of Image for one Touch Assessment
Numericalization Of Brightness of Image for one Touch Assessment
Our software makes it possible to see differences in fluorescence that are hard
to judge
with the naked eye
We explored a way to detect the fluorescence obtained by the RT-LAMP reaction described above.
Our
software enables detection which is a key part of the DLA cycle.
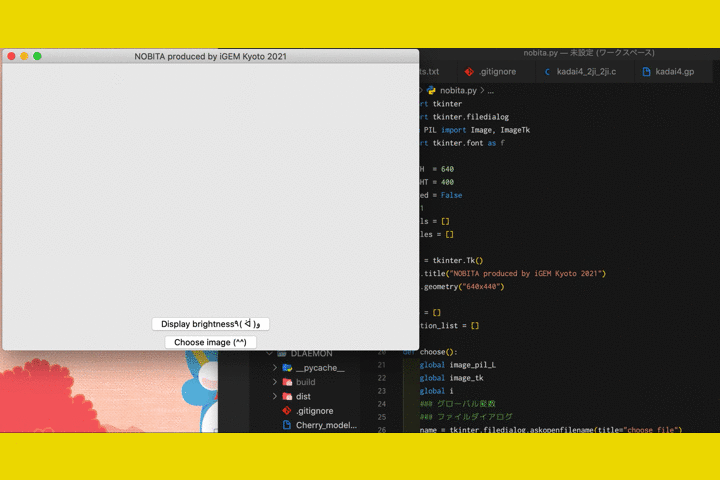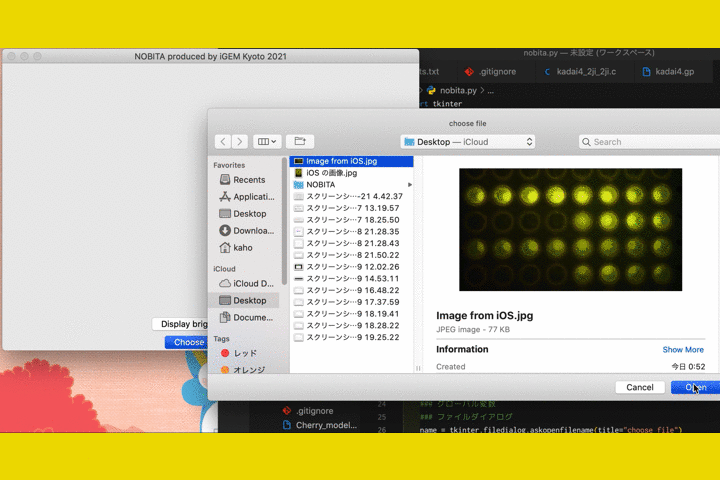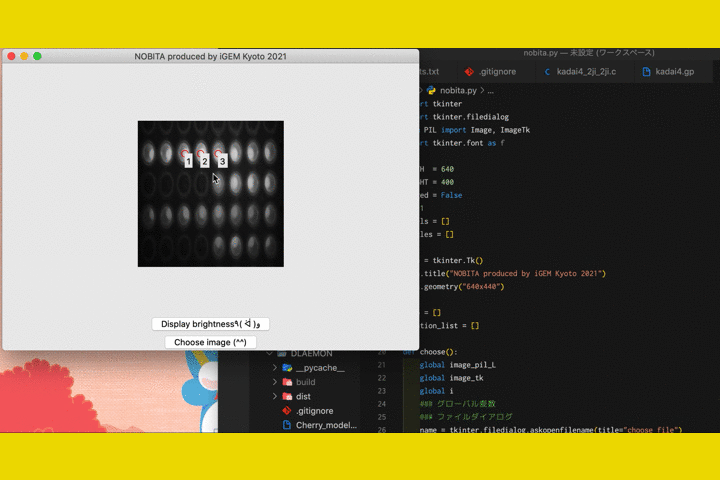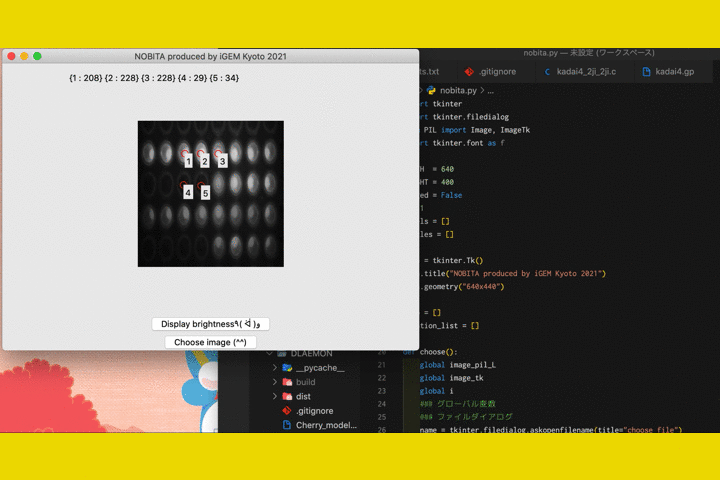
Fig.9 The screen of fluorescence detection
By numericizing the fluorescence, it becomes clearer to determine the presence or absence of fluorescence in photos taken with hardware.
By numericizing the fluorescence, it becomes clearer to determine the presence or absence of fluorescence in photos taken with hardware.
Judging the existence of fluorescence is often challenging for a person. However, if there is
software
that
can quantify fluorescence, it is expected that the evaluation will become simpler for everyone.
Through
the
development and verification of NOBITA, we have demonstrated that fluorescence can be reliably
determined.
5. DLA cycle
DLA is a system that "combines both diagnostic imaging and nucleic acid detection, compensating
for the
weaknesses of each component to enable more convenient and sophisticated virus detection".
As mentioned above, we proved that we have developed and created an enzyme reaction, hardware, and software with the expected performance for all the steps as a unified project.
As mentioned above, we proved that we have developed and created an enzyme reaction, hardware, and software with the expected performance for all the steps as a unified project.
This DLA Cycle integrates the elements necessary for diagnosis. That is why the results obtained
in each
element feedback on each other to advance machine learning and increase the accuracy of the
DLAEMON
determination. Furthermore, there are merits of unified development. One is the accumulation of
image data
with unfavorable evaluations whose results from two components do not agree. The smarter DLAEMON
becomes,
the more it may be able to reject such images even in advance that the results of image judgment
and
nucleic
acid detection do not agree with each other. It may even tell us the appropriate photographing
technique
to
obtain an accurate diagnosis. Sooner or later, DLAEMON alone may suffice to make a diagnosis.
Killing Pests
- in vivo produced dsRNA effectively kills pests by RNA
interference
The control of thrips by RNAi has already been reported[1]. However, in
this case, dsRNA was synthesized
in
vitro at great cost. For future application as a spray-type pesticide, it is required to prepare
a large
amount of dsRNA solution, and the conventional synthesis method is not suitable for this
purpose.
Therefore,
we attempted to produce the previously reported dsRNA by synthesis in E. coli at a reduced
cost.
Unfortunately, production in E. coli has the problem that the content of the dsRNA obtained solution is impure. Thus, we verified whether the dsRNA solution obtained by our method could still be effective for RNAi.
We conducted the assay in the same manner to the reported case[1]. The results showed that the dsRNA we synthesized in E. coli contributed to the increased mortality of thrips on leaves soaked in the dsRNA solution. This proves that our dsRNA pesticide can be an alternative to conventional pest control.
Unfortunately, production in E. coli has the problem that the content of the dsRNA obtained solution is impure. Thus, we verified whether the dsRNA solution obtained by our method could still be effective for RNAi.
We conducted the assay in the same manner to the reported case[1]. The results showed that the dsRNA we synthesized in E. coli contributed to the increased mortality of thrips on leaves soaked in the dsRNA solution. This proves that our dsRNA pesticide can be an alternative to conventional pest control.

Fig.10 The graph of the death rate of thrips
Since the death rate of thrips on leaves which absorbed dsRNA was higher compared with that of the negative control, the result reflects the effects of RNAi induced by our dsRNA.
Since the death rate of thrips on leaves which absorbed dsRNA was higher compared with that of the negative control, the result reflects the effects of RNAi induced by our dsRNA.
In this experiment, we used the same protocol as one already reported in the paper in which we
indirectly
fed thrips dsRNA via a leaf soaked in the solution. The results of the elimination with this
method should
lead to the realization of the application of general and convenient methods of pesticide usage,
such as
spraying, only if sufficient amounts of dsRNA are attached to the leaves. The result that it is
possible
to
produce large amounts of dsRNA with E. coli will also support this spraying method which
requires large
amounts of dsRNA (see the Engineering
page).
BLOOM
1. Simulation by mathematical modeling predicts the behavior of the BLOOM
system
Our BLOOM system works based on the interaction of two plasmids and proteins that they express.
It is
regulated by multiple steps of reactions by each of the components, and the behavior of the
whole system
is
considered to be ruled by multiple parameters. In order to determine the key factors prior to
conducting
lengthy and expensive wet lab experiments, we examined the behavior of the system in
silico by taking
advantage of the mathematical modeling.
To begin with, we tried to confirm that the plasmid D is not inherited due to its asymmetric
distribution.

Fig.11 Simulation of plasmid D loss
The simulation shows plasmid D is eventually lost from almost all of the cells.
The simulation shows plasmid D is eventually lost from almost all of the cells.
Next, we aimed to demonstrate that the repressors expressed from plasmid D are gradually lost
from the
cells
following its drop-out, so that the expression of two reporters are de-repressed in a stepwise
manner.

Fig.12 Two reporters expressed with a time difference
The simulation shows the two reporters are expressed with a time interval in between, following the loss of plasmid D.
The simulation shows the two reporters are expressed with a time interval in between, following the loss of plasmid D.
These simulations prove that the BLOOM system would work as desired if each parameter could be
replicated
according to the conditions of our model. Moreover, it suggests that any genes of interest
substituting
the
two reporters could be expressed with a temporal difference.
2. Engineered ssrA tag derivatives with various
protein degradation efficiencies
Nowadays, types of biomolecules produced for research or industrial uses are highly
wide-ranging, and so
are
the methods of their production. To adapt our BLOOM system to these various needs we focused on
controlling
the time interval between the expression of two genes.
Hence, we used modeling to determine what factor has the most influence on the time difference
of gene
expression. As a result, we found that among the parameters such as the degradation rate of the
repressors,
the strength of each promoter driving the expression of each repressor, and the copy number of
the
plasmid,
the degradation rate of the repressor was shown to most critically contribute to the length of
the time
difference.

Fig.13 Protein degradation rate of the repressors is the key parameter
of the length
of expression interval
This result suggests that we would be able to flexibly regulate the time difference by controlling the degradation rate of repressors. We focused on developing methods to regulate protein degradation (see the Model page).
This result suggests that we would be able to flexibly regulate the time difference by controlling the degradation rate of repressors. We focused on developing methods to regulate protein degradation (see the Model page).
Peptide tags that promote the degradation of fusion proteins are generally used to control
protein
half-life. SsrA tag is reported to efficiently causefusion protein degradation through the
interaction
with
cellular proteases, and several mutants with differing activities have been identified[2]. However, the
repertoire of tags is still not large enough to enable a flexible control of protein half-life.
Therefore,
we aimed to obtain a collection of mutant tags with various degradation efficiencies by
mutagenizing the
wildtype ssrA tag. We fused the ssrA tag sequence to GFP while introducing mutations in the tag
by
random-base primers, and cloned the mutant library of ssrA-tagged GFP into a plasmid vector, so
that
mutant
tags of different activity can be identified by comparing GFP intensity of E.coli transformants.

Fig.14 E.coli cultures with various GFP intensity
As a result, we successfully isolated E.coli clones with various GFP intensity, and identified 17 mutant ssrA tags which would be able to be used to regulate protein degradation rate by 17 different levels (see ssrA results in the Results page).
As a result, we successfully isolated E.coli clones with various GFP intensity, and identified 17 mutant ssrA tags which would be able to be used to regulate protein degradation rate by 17 different levels (see ssrA results in the Results page).

Fig.15 Comparison of GFP intensity between mutant tags
The fluorescence of each culture was quantified by ImageJ as inverted mean gray value. Measured value of each culture was then normalized to the level of the culture expressing non ssrA-tagged GFP (no ssrA tag), and presented as mean ± SD from three biological replicates. WT (LAA) and two mutants reported in a previous research are shown in red boxes.
The fluorescence of each culture was quantified by ImageJ as inverted mean gray value. Measured value of each culture was then normalized to the level of the culture expressing non ssrA-tagged GFP (no ssrA tag), and presented as mean ± SD from three biological replicates. WT (LAA) and two mutants reported in a previous research are shown in red boxes.
These results demonstrate that the mutant tags that we identified enable more precise control of
the
repressors by freely changing the tag sequence. Furthermore, it suggests that based on our
method, it would be possible to identify even more mutant tags to expand the repertoire. These
results should prove useful for fine-tuning our BLOOM system.
References
- Andongma, A.A., Greig, C., Dyson, P.J., Flynn, N., and Whitten, M.M.A. (2020) "Optimization of dietary RNA interference delivery to western flower thrips Frankliniella occidentalis and onion thrips Thrips tabaci", Arch. Insect Biochem. Physiol. 103, e21645.
- Flynn, J.M., Levchenko, I., Seidel, M., Wickner, S.H., Sauer, R.T., and Baker, T.A. (2001) "Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis", Proc. Natl. Acad. Sci. U. S. A. 98, 10584–10589.
