RDX Genetic Circuit
Device #1: Detection of RDX (BBa_K3670001)
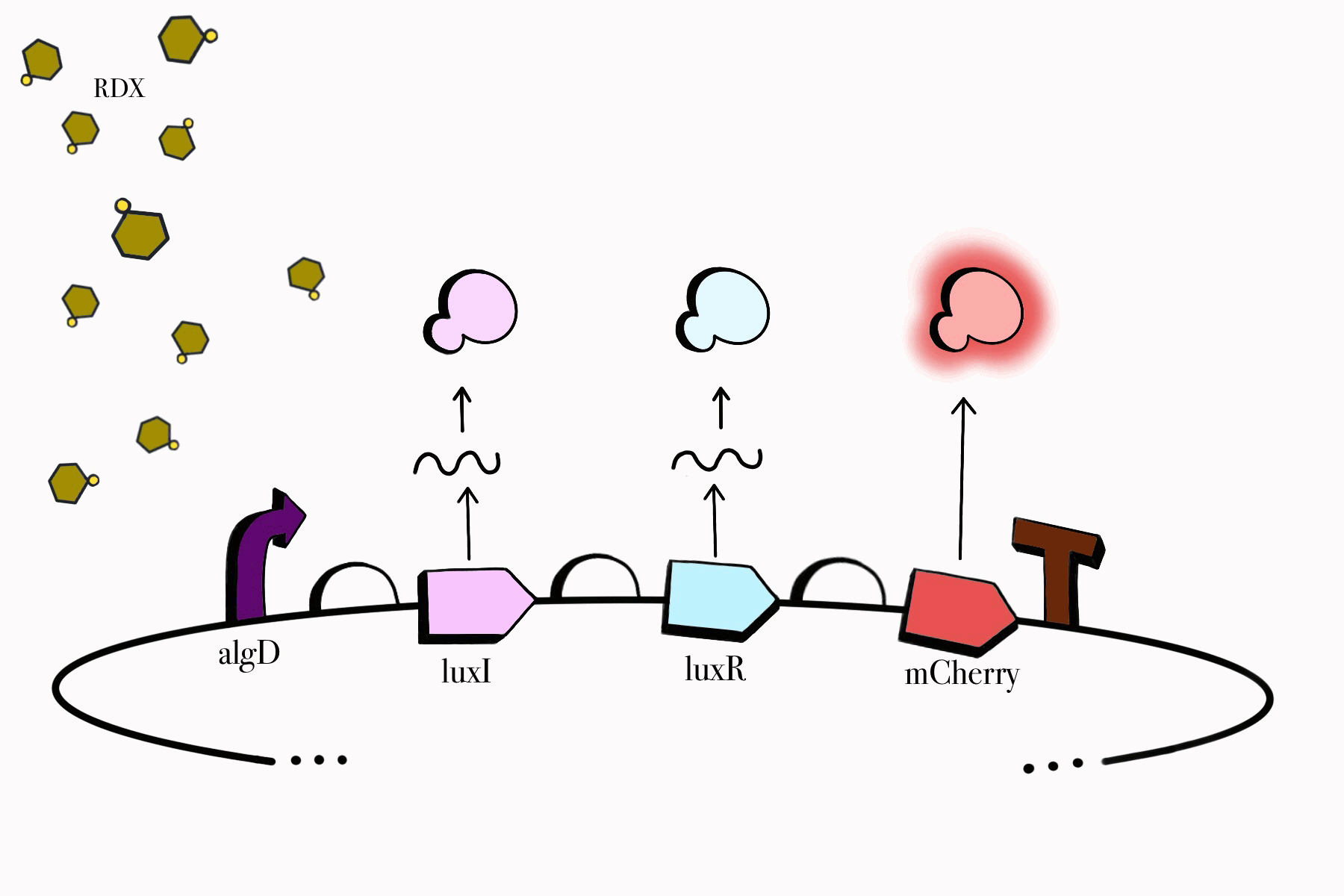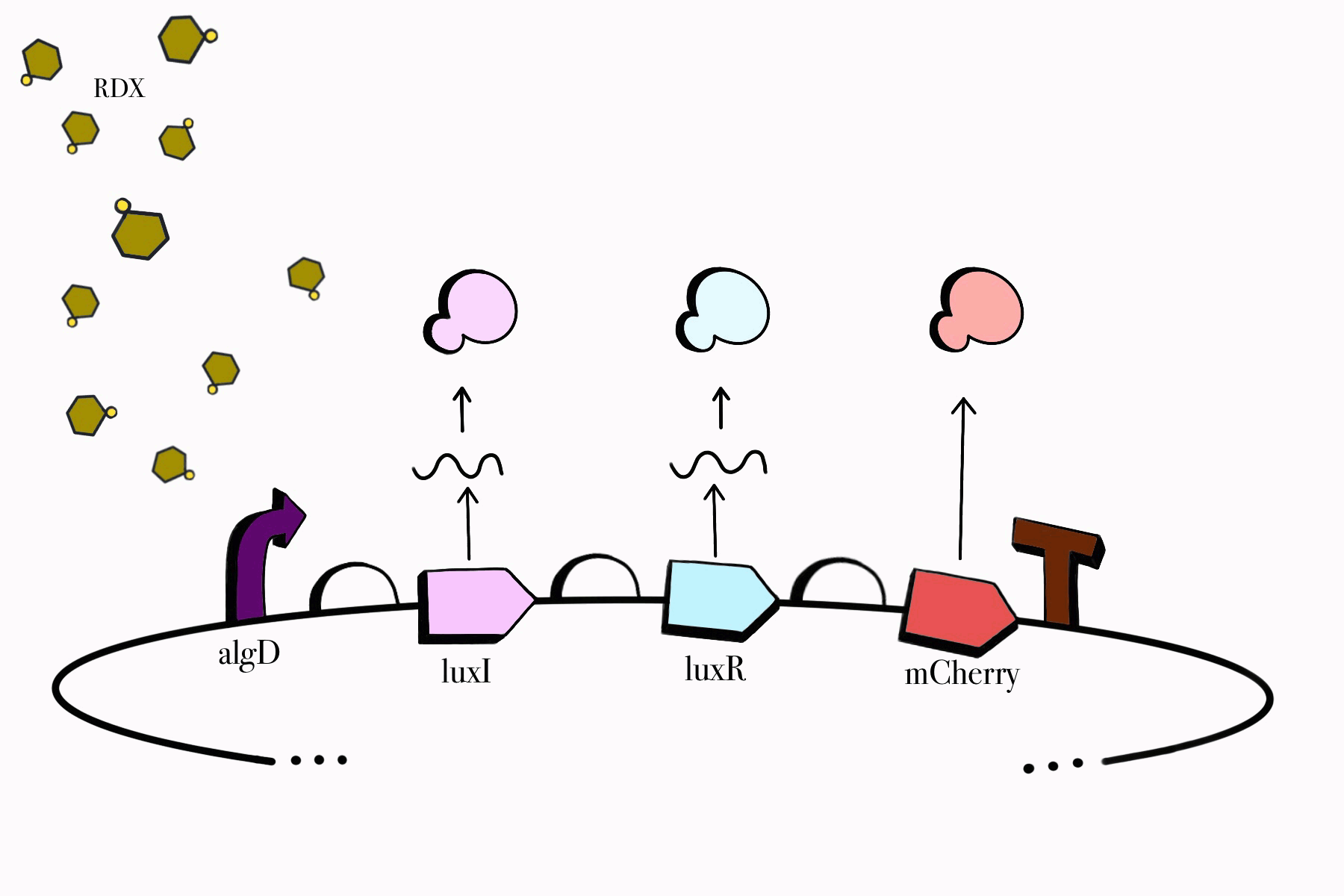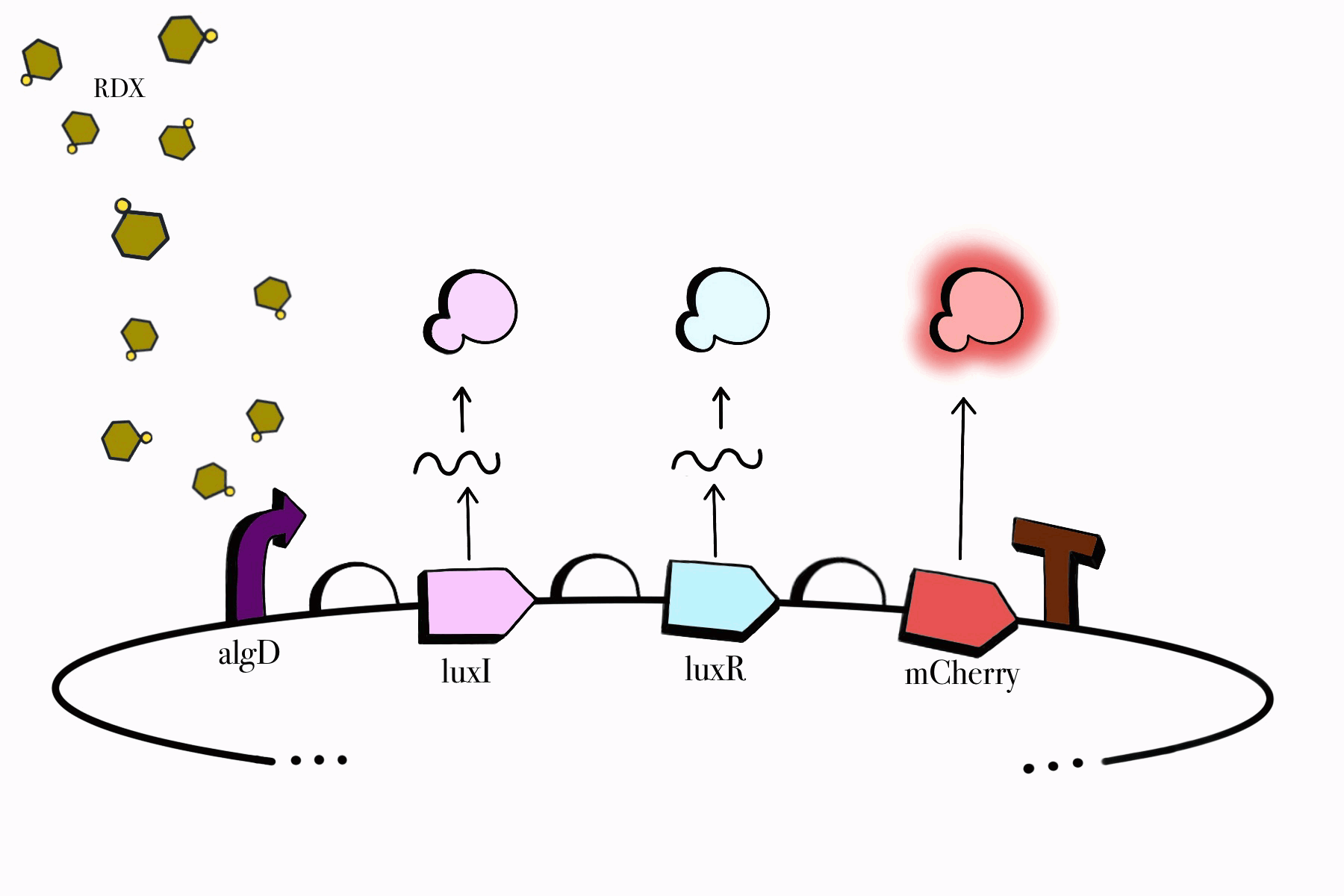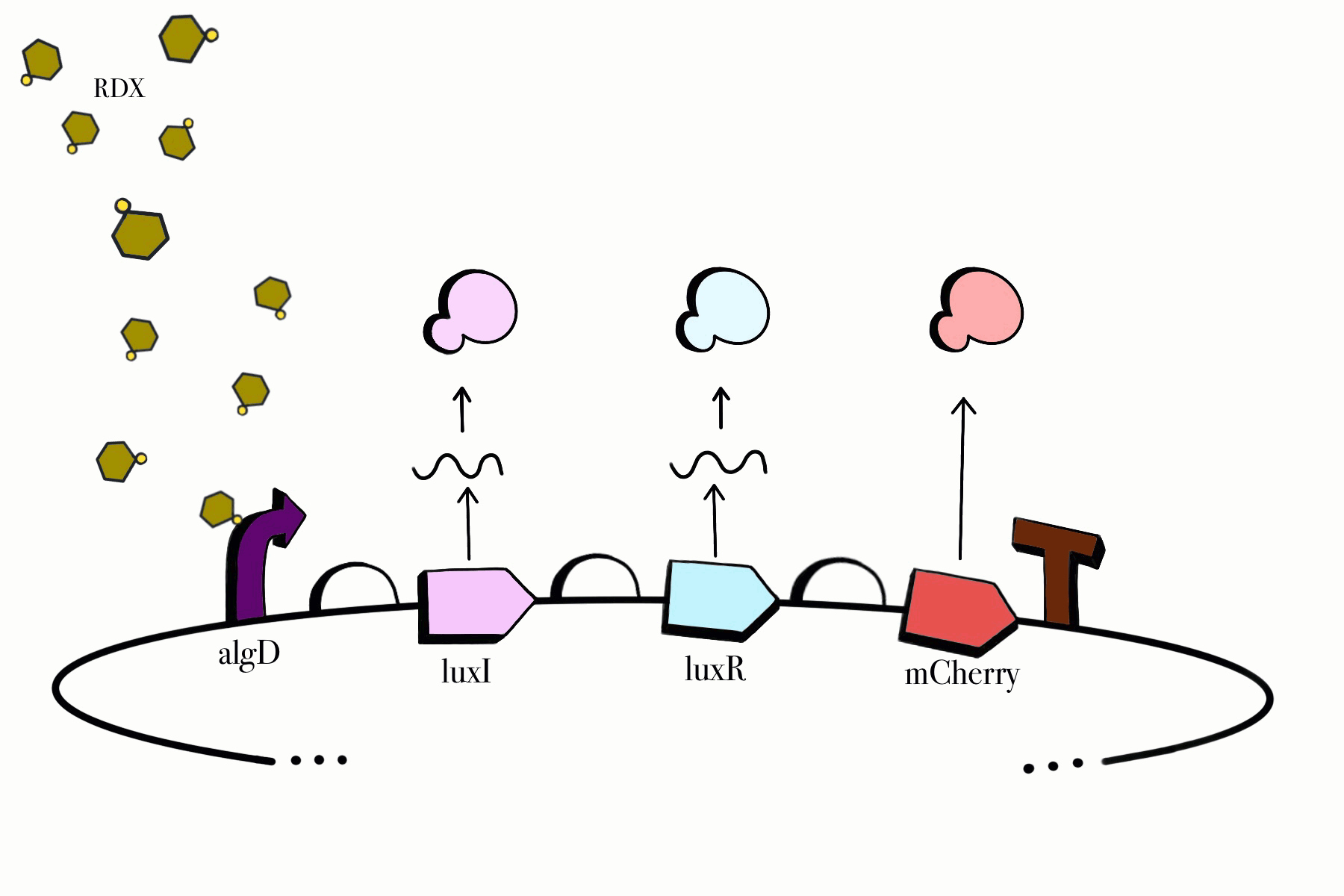
For the first device, the promoter algD is induced by a certain range of concentration (between 0.2-0.5mmolar) of RDX. The sequence of the algD promoter and its regulators were derived from analogous sequences from the genome of the Pseudomonas species. For this cycle, we revised and adjusted the original sequence, which was lacking certain regions that promotes the regulation of the promoter algD. Following the promoter are the luxI and luxR genes. These genes will enable the chassis to partake in a quorum sensing system with the second device, which will enable them to act in accordance to the RDX concentrations in their environment. The product of luxI will convert S-adenosylmethionine (SAM) into acyl-homoserine lactone (AHL). Consequently, luxR will produce a protein which binds to AHL. This merge will stimulate the transcription of luxpr (pLuxR) promoter in the second device. To visualize the transcription of this device, the reporter gene mCherry was included, which produces a red fluorescence indicating that gene transcription is operating. Lastly, this sequence ends with the BioBrick terminator.
| Part | Function and Usage |
|---|---|
| algDpromoter BBa_K3857000 | Transcription of the algD promoter is induced as a stress response towards the presence of RDX. The main reason for using an inducible promoter is to assure that the activity occurs only in the presence of RDX, as well as to quantify the activities o the device at different concentrations. By using this promoter, we look to create a bio sensing system and be able to regulate future biodegradation. |
| LuxI BBa_C0061 | LuxI is a synthase that converts S-adenosylmethionine (SAM) into acyl-homoserine lactone (AHL), which can diffuse across cell membranes. AHL signal molecules are recognized by LuxR-type receptors, which constitute a class of transcription factors that possess an amino-terminal AHL binding domain and a carboy-terminal DNA binding domain (Li and Nair, 2012). |
| LuxR BBa_C0062 | When LuxR is bound to AHL, it produces a protein that stimulates transcription of the right hand lux promoter, luxpr. The expression of the AHL synthase LuxI is also unregulated by LuxR, resulting in a positive feedback loop. An increased production of AHL and LuxR as a result of RDX would increase the transcription response in Device 2. | mCherry BBa_J176005 | In the presence of RDX, the reporter gene emission of the red fluorescence protein can be quantified to analyze the transcription of the first device. Through the usage of fluorometry, the activity of the first device is measured between a correlation of the intensity of color emission while measuring the concentrations of RDX. The mCherry gene expresses a red light that has emission at 610nm and excitation at 597nm. | Terminator | Our terminator is a sequence inserted to halt the transcription of our first device, based on the BioBrick terminator. |
Device #2: Biodegradation of RDX (BBa_K3857003)

Activation of our second device for Biodegradation will be governed by the presences of LuxR bound to AHL, which will act as the inducer for this endeavor. As such, prlux will act as the promoter for this device. When activated, gene expression of xplB and xpla will take place. The xplB gene encodes for a partner flavodoxin reductase, while the xplA encodes for flavodoxin domain fused (at the N-terminus) of a P450 cytochrome. The product of these genes will create a system in charge of the denitrification of RDX taking two possible routes: anaerobic or aerobic pathways, followed by subsequent ring cleavage of RDX. This ring cleavage is essential to produce formaldehyde and nitrite, our secondary products essential for the activation of our third device. When biodegradation of RDX is complete, the gene amilGFP, a yellow fluorescent protein, will function as a reporter gene.
| Part | Function and Usage |
|---|---|
| luxpR BBa_R0062 | This promoter is key to producing our proteins of interest: xplA and xplB. It is unregulated by the activation of the LuxR activator protein which forms a complex with autoinducer AHL. The addition of this promoter contributes to our efforts of quorum-sensing and a regulated high transcription rate. |
| xplA BBa_K3670004 | Encodes for flavodoxin domain fused (at the N-terminus) of a P450 cytochrome. Studies demonstrate unequivocally that his gene product is necessary for RDX degradation (Chong et al. 2014), especially in the cases where bacterial species do not have natural RDX-degrading systems. xplA, along with xplB, work together as a system to catalyze reductive denigration of RDX and subsequent ring cleavage, both under aerobic and anaerobic conditions (Chong et al., 2014). |
| xplB BBa_K3857002 | This gene encodes for xplA partner, a flavodoxin reductase. This reductase is dependent on NADPH and has FAD as a cofactor. XplB is involved in the activation of the catalytic center of XplA via electron transfer from NADPH to a flavodoxin domain fused to the N-terminal of the P450 domain of xplA (Sabir, Grosjean and Bruce, 2017). Even though it is not required, XplB contributes to RDX catabolism, reinforcing the already efficient activity of XplA. Some bacteria have shown that the absence of xplB (with an existing xplA in the system) reduced the rate of RDX degradation by 70% (Chong et al, 2014). |
| amilGFP with degradation LVA tag BBa_K592010 | Involved in the expression of green fluorescence protein, as well, encodes for a small peptide functioning as a degradation tag that will allow for fine-tuning protein levels and thus regulating of the GFP in the bacteria. | Terminator | Our terminator is a sequence inserted to halt the transcription of our first device, based on the BioBrick terminator. |
Device #3: Killswitch of RDX Circuit

The third device, considered as a killswitch, is a modified AND gate that is activated by the presence of formaldehyde and nitrite. The presence of formaldehyde activates the pfirm promoter. The pfirm promoter activation then allows the transcription of the supD gene, while the presence of nitrite activates the inducible promoter pyeaR, which allows the transcription of the t7ptag gene. Each promoter possesses a level of regulation due to the nsrr binding sequence. NsrR protein will bind to this specific sequence to repress transcription. Such a sequence is native to the pyeaR. However, due to basal expression detected by NEU_China 2019, an extra binding sequence of nsrr was added after the promoter. In addition, we decided to add this sequence downstream of pfirm to add more regulation to the killswitch. The t7ptag gene codes for a polymerase with two amber or nonsense mutations which inhibits its translation. To modify the amber mutations, the supD gene produces a tRNA that suppresses them. This then activates the P17 promoter and allows expression of the colicin gene. Colicin will cause bacterial lysis, allowing the detection and biodegradation of RDX to cease, and the bacteria to die, promoting biosafety.
| Part | Function and Usage |
|---|---|
| Pyear promoter BBa_K216005 | This inducible promoter will be activated in the presence of nitrate, nitric oxide or nitrite. Once nitrite and nitrate enter Escherichia coli, they will be converted into nitro oxide It is important that transformation to nitric oxide occurs so that this promoter can be inactivated and transcription of unwanted genes does not proceed. PyeaR works successfully in both anaerobic and aerobic conditions. |
| Formaldehyde-Inducible Promoter pfirm BBa_K2728001 | Inducible promoter that will be activated in the presence of formaldehyde. |
| SupD + terminator BBa_K228100 | SupD produces a tRNA amber mutation suppressor that activates the mRNA produced by t7ptag. |
| T7ptag (T7polymerase with amber mutation) BBa_K228000 | This is a gene that encodes for a T7 polymerase with two amber mutations. For a successful translation of this gene’s mRNA to take place, an Amber mutation suppressor, SupD, must be available to eliminate these mutations. T7ptag, as well as supD, is what makes the third device function as an AND gate. |
| PT7 BBa_K2406020 | This promoter activates in presence of t7ptag. Activation of this gene expression will result in production of colicin. |
| Lysis gene BBa_K117000 | The lysis gene encodes for the lysis-protein in bacteria that produce colicin. It activates the endonuclease activity of colicin, killing our bacteria after biodegrading RDX. |
References
Chong CS, Sabir DK, Lorenz A, Bontemps C, Andeer P, Stahl DA, Strand SE, Rylott EL, Bruce NC. Analysis of the xplAB-containing gene cluster involved in the bacterial degradation of the explosive hexahydro-1,3,5-trinitro-1,3,5-triazine.Appl Environ Microbiol. 2014 Nov;80(21):6601-10. doi: 10.1128/AEM.01818-14. Epub 2014 Aug 15. PMID: 25128343; PMCID: PMC4249041.
Jackson, R. G., Rylott, E. L., Fournier, D., Hawari, J., & Bruce, N. C. (2007). Exploring the biochemical properties and remediation applications of the unusual explosive-degrading P450 system XplA/B. Proceedings of the National Academy of Sciences, 104(43), 16822-16827. doi:10.1073/pnas.0705110104
Lee, B., Baek, H., & Oh, K. (2013). Use of an algD Promoter-Driven Expression System for the Degradation of Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by Pseudomonas sp. HK-6. Current Microbiology, 67(4), 480-486. doi:10.1007/s00284-013-0387-5
Li, Z., Nair, S.K. (2012). Quorum sensing: how bacteria can coordinate activity and synchronize their response to external signals. Protein science: a publication of the Protein Society, 21(10), 1403-1417. https://doi.org/10.1002/pro.2132
Sabir DK, Grosjean N, Rylott EL, Bruce NC. Investigating differences in the ability of XplA/B-containing bacteria to degrade explosive hexahedron-1,3,5-trinitro-1,3,5-triazine (RDX). FEMS Micriobiol Lett. 2017 Aug 1;364(14). don: 10.1093/femsle/fnx144. PMID: 28854671.
