1. Research
Our aim with this project is to find a modern solution to a problem that has been plaguing the citizens of Vieques for several decades. As we all know, the small puertorrican municipality has been dealing with contamination issues due to the bomb detonations that occurred near its several bodies of water, one of these being the Anones Lagoon in Vieques. Several areas of the municipality were used for military training, and were recently declared as a Superfund site (Sanderson et al., 2017).
According to Sanderson et al. 2017, the human health risks that come from exposure to the contaminants brought by the bombs aren't being controlled. Contamination has been detected at several sites at very concerning levels to the point where it is considered unsafe to thread near those areas, and exposure to this contamination might lead to unexpected health complications. Due to this, we decided on finding a possible solution to this problem throughout the use of Synthetic Biology.
2. Design
Out of all the contaminants present in Vieques, we chose hexahydro-1,3,5-trinitro-s-triazine (RDX) to be the focus of our project. Rosen et al. (2017) held a study to analyze the concentration of RDX and other explosives in the waters in the municipality. Chakraborty et al. 2008 also spoke about how the leaching of explosive compounds, such as RDX, occurs due to unexploded munitions present in the area.
The XplA/XplB gene cluster present in our genetic circuit was first found in Rhodococcus sp., a bacterium with the potential of fulfilling the biodegradation of RDX in aerobic environments (Bernstein et al., 2011). We believe that these two genes will enable us to conceptualize and eventually construct in vitro a bacterial prototype capable of fulfilling this biological degradation of RDX, to serve as a possible solution to the contamination problems found in Vieques.
3. Build
Several publications were used as a reference for the analysis and characterization of the XplA/XplB gene cluster (Jackson et al., 2007; Halasz et al., 2012; Cary et al., 2021). Once we obtained the genetic sequence, we started crafting our genetic circuit using SnapGene and Benchling to facilitate the visualization of our genetic devices. This helped us visualize our cloning workflow and choose the restrictions sites to permit the synthesis of our genetic constructs.

Figure 1. Cloning workflow for the assembly of Device 1 of our RDX Biodegradation Genetic Circuit, built using SnapGene.
We made sure to build our genetic devices in such a way that would enable us to easily detect their functionality, while simultaneously keeping them under control and only functioning when specific conditions are met. Reporter genes, fluorescent proteins in our case, would aid us in detecting them as our cloned bacteria grows in culture media. Our chosen promoters also ensure that our devices only work in the presence of RDX and their assigned compounds, meaning our genetic circuit won't be able to jumpstart itself unless it is found in the desired environmental conditions. Lastly, our third device contains a killswitch which will serve as a biocontrol to prevent unnecessary cellular proliferation in the environment, enabling cell lysis once our cloned bacteria fulfills its function.
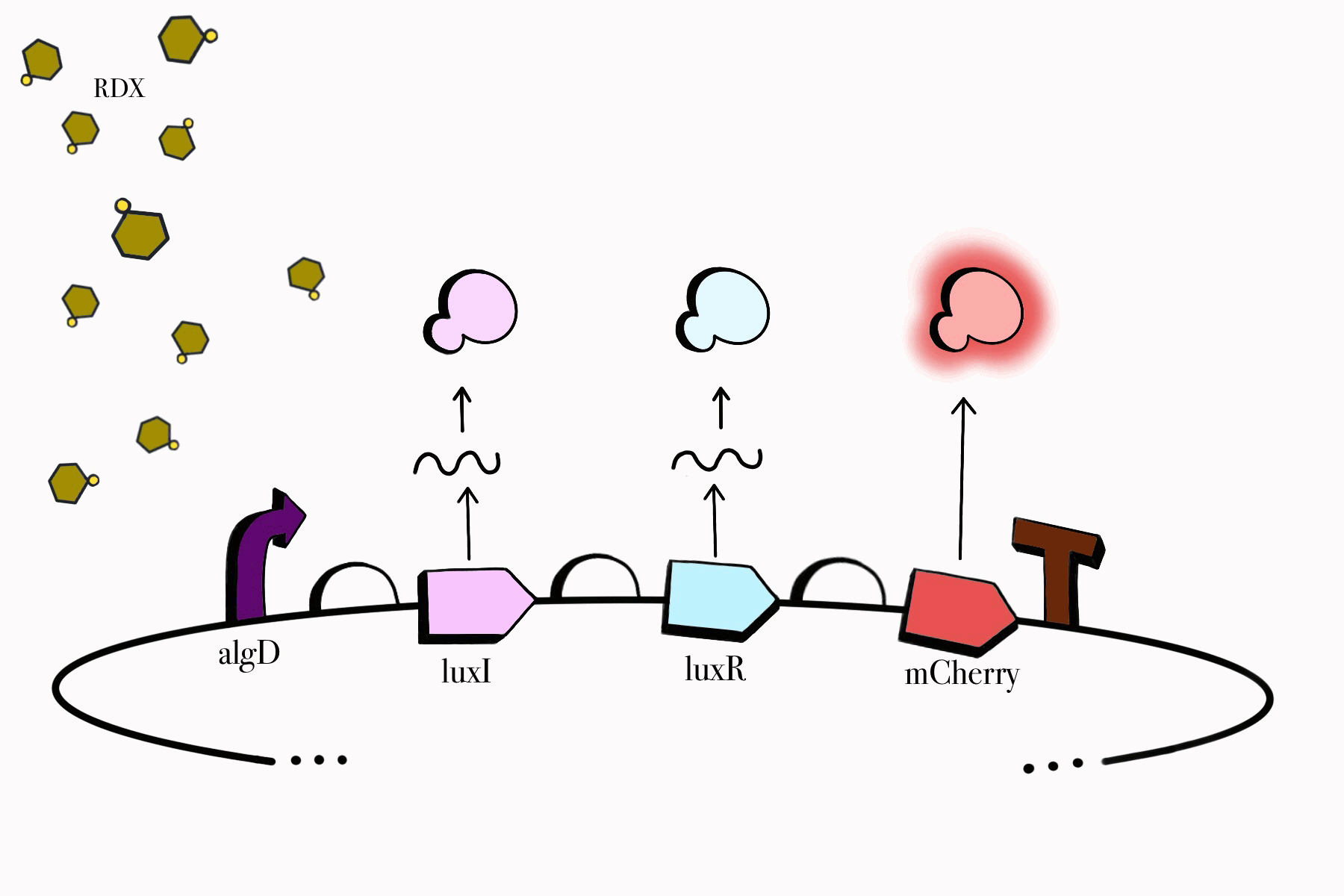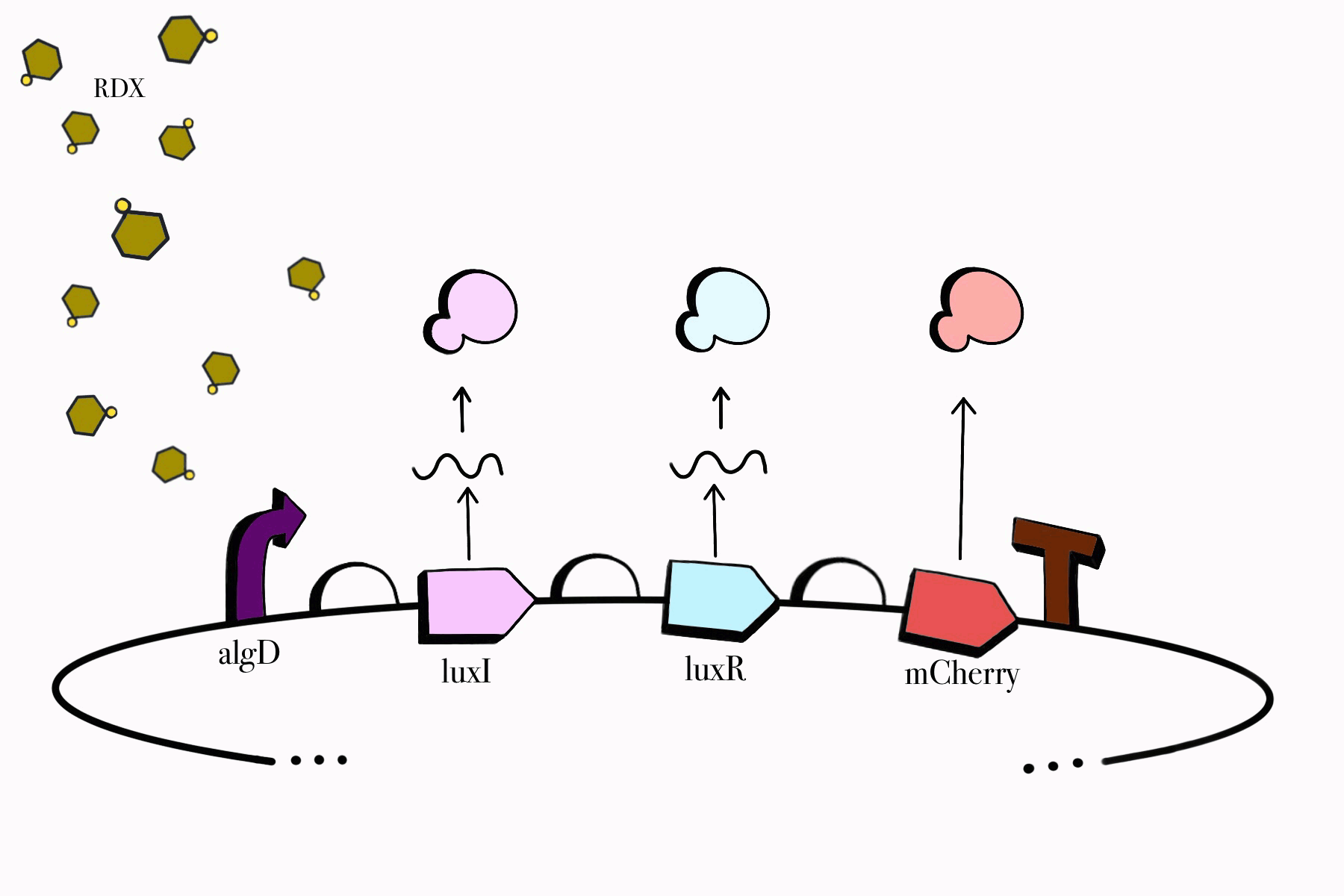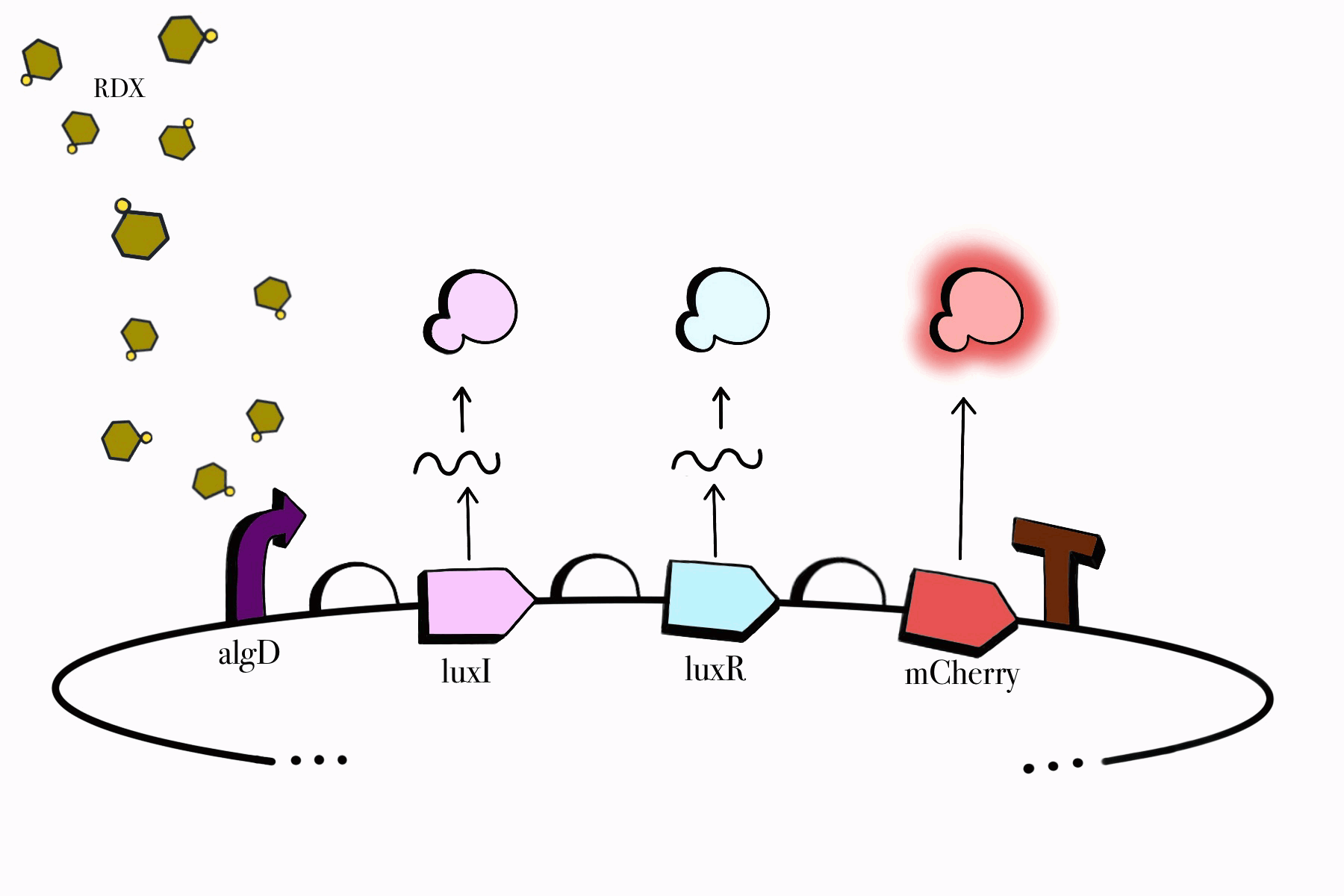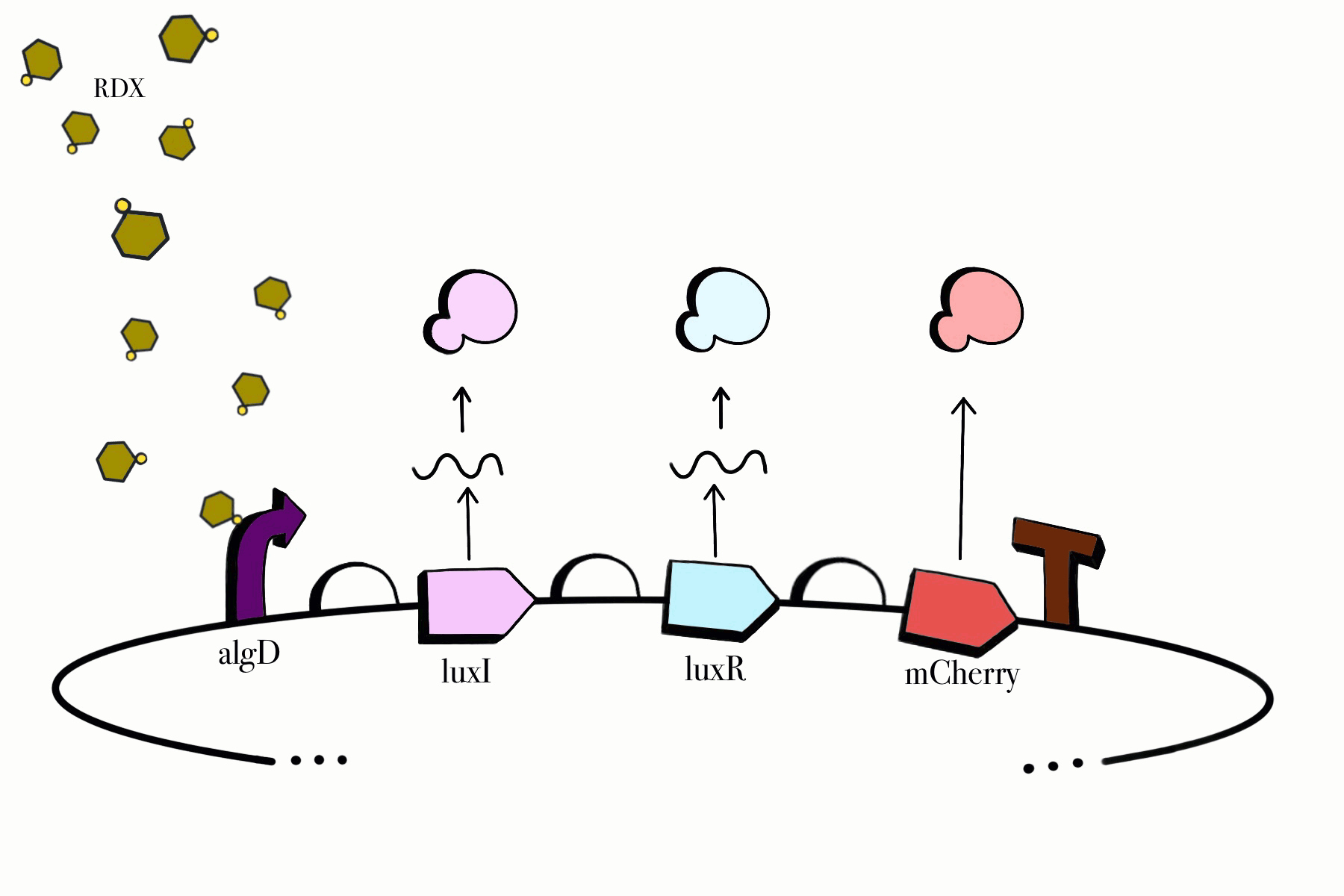
Figure 2. SBOL representation of the RDX Device #1: Detection
4. Test
Our first attempts to clone our DNA fragments were made using the Linearized Plasmid Backbones sent out in the 2019 iGEM Distribution kits. The three different plasmids present in the kit allowed us to prepare a 3A cloning workflow which would allow us to prepare stocks of our two fragments in two different plasmids and a single plasmid with both of the needed fragments inside of it. Several weeks later we decided on taking a different approach to our cloning techniques and switched our plasmid to pUC19. Each fragment was cloned individually on its own pUC19 plasmid. Lastly, we expect to validate our devices using fluorometric assays in a 96-well plate reader. More information regarding our proof of concept can be found here.
5. Learn
Our original cloning plans could not be fulfilled due to several unforeseen circumstances that appeared while attempting to transform our bacteria to express our plasmids containing our desired DNA fragments. Pink, fluorescent colonies were observed, even on bacterial colonies belonging to plates that didn't contain the pink fluorescent protein gene. We held additional contamination assays to see what exactly happened and why we were obtaining these unexpected results. Our final decision was to change the plasmids we were using, which is why we resorted to working with pUC19 instead to fulfill our cloning strategy.
6. References
Cary, T. J., Rylott, E. L., Zhang, L., Routsong, R. M., Palazzo, A. J., Strand, S. E., & Bruce, N. C. (2021). Field trial demonstrating phytoremediation of the military explosive RDX by XplA/XplB-expressing switchgrass. Nature Biotechnology, 1-4.
Chakraborty, R., Ramos-Hernandez, N., Perez, E. X., Katsuura, Y., Geller, J. T., Massol-Deya, A., & Hazen, T. C. (2008). Characterization of novel marine sulfate-reducing bacteria resistant to RDX and other explosives.
Halasz, A., Manno, D., Perreault, N. N., Sabbadin, F., Bruce, N. C., & Hawari, J. (2012). Biodegradation of RDX nitroso products MNX and TNX by cytochrome P450 XplA. Environmental science & technology, 46(13), 7245-7251.
Jackson, R. G., Rylott, E. L., Fournier, D., Hawari, J., & Bruce, N. C. (2007). Exploring the biochemical properties and remediation applications of the unusual explosive-degrading P450 system XplA/B. Proceedings of the National Academy of Sciences, 104(43), 16822-16827.
Rosen, G., Colven, M., George, R., Lotufo, G., Woodley, C., Smith, D., & Belden, J. (2017). Validation of passive sampling devices for monitoring of munitions constituents in underwater environments. Space and Naval Warfare Systems Command San Diego United States.
Sanderson, H., Fauser, P., Stauber, R. S., Christensen, J., Løfstrøm, P., & Becker, T. (2017). Civilian exposure to munitions-specific carcinogens and resulting cancer risks for civilians on the Puerto Rican island of Vieques following military exercises from 1947 to 1998. Global Security: Health, Science and Policy, 2(1), 40-61.
