Overview
As of now, there is very little research on RDX biodegradation that provides a cost-effective solution on land and water contamination. In addition, the algD promoter is a rather novel sequence that is getting more attention due to its affinity to RDX (referencia). Thus, we sought to create complementary genetical devices that allow for detection and degradation of the nitroexplosive using palgD as an inducible promoter. To characterize these novel devices, we implemented gene reporters to quantify the intensity of said devices. As such, the emission of mCherry will tell us if the device is working as desired; the second device, fitted with amilGFP, will tell us the functionality of the gene expression, but not the degradation of RDX. In hypothetical terms, to know if RDX is being degraded by the xplA/B protein, we will utilize fluorometric assay, as stated in our work plan, to measure each device separately and then all the devices together.
One important aspect of the degradation is that it can occur under aerobic or anaerobic conditions (Sabir et. al., 2017). From any of these two conditions, nitrite and formaldehyde are produced as byproducts from the degradation. These substances are used as inducers for our third device, the killswitch. However, these substances are considered toxic (Liteplo et al., 2002; Thompson, 2021). To ensure biosafety, stirred batch bioreactor system was developed to maintain a level of regulation of these substances contained in the system.
Originally, we decided to use Pseudomonas putida KT2440 as our preferred chassis since it is considered to be one of the most preferred environmental organism that has activity against aromatic pollutants (Belda et al., 2016). In addition, its complex metabolism allows P. putida to have an impact on biotechnological use for environmental cleanup (Belda et al., 2016). However, we decided to utilize NEB 5-alpha competent E. coli cells and NEB 10-beta Competent E. coli, which are derived from E. coli Dh5 alpha and E. coli Dh10B, respectively. Each contains endA1 mutation, allowing for plasmid preparation. This would permit us to effectively characterize the functionality of our devices to later implement them into P. putida KT2440, since it would be, in theory, a practically better chassis for the environment it would be put in. Thus, each device underwent a codon optimization for E. coli Dh5alpha and Dh10B strains.
Regarding our chassis, we were concerned about the workload that the bacteria would have to put in with all three devices expressing in the same organism. Thus, we implemented luxI and luxR to allow quorum sensing to take place and provide a means of communication between bacterias in response to RDX. In this case, this phenomenon involves the acyl-homoserine lactone (acyl-HSL), which is produced by the luxI synthase, and luxR proteins, which mediates the response and complexes with acyl-HSL(Callahan & Dunlap, 2000; Li & Nair, 2012). Here, we decided to put Device 1 and Device 3 separate from Device 2 and Device 3 in a plasmid. Theoretically, the AHL/LuxR complex will allow bacteria to produce the xplA and xplB proteins in accordance with the level of concentration of RDX.

Figure 1. Two E. coli Dh5 alpha with Device 1 and 3 (on the left side) and Device 2 and 3 (on the right side). The colored bacteria represents the color that will show if gene expression has taken place (red is due to mCherry while yellow is due to amilGFP). The red bacteria produces AHL and LuxR complex to induce gene expression of the xplA and xplB genes in the yellow bacteria.
RDX Genetic Circuit
The RDX system is composed of three genetic devices, each with a specific function: detection, biodegradation, and lysis, which will be regulated by quorum sensing. This device begins with the stress sensitive promoter algD, which will initiate transcription in the presence of RDX. Later, LuxI gene will create a synthase capable of creating acyl-homoserine lactones (AHL) that will bind to LuxR protein. The binding of these two molecules creates a transcription factor that will activate LuxpR promoter. It will then begin the transcription of the second device, that will contain the xplA and xplB genes, which produces enzymes capable of degrading RDX. After transcription of xplA and xplB genes has been completed, the amilGFP gene will be transcribed. GFP allows us to identify whether the enzymes are being produced by emitting a yellow fluorescence. The end-products of RDX, specifically nitrite and formaldehyde, will act as transcription factors in the third device, which is the killswitch. Lastly, the kill switch circuit will be controlled by a modified synthetic-AND Gate, which will allow bacterial lysis by requiring the presence of the byproducts of the biodegradation of RDX: formaldehyde and nitrite. Lysis will initiate due to the presence of colicin and, therefore, stop bacterial transcription.
Device #1: Detection of RDX
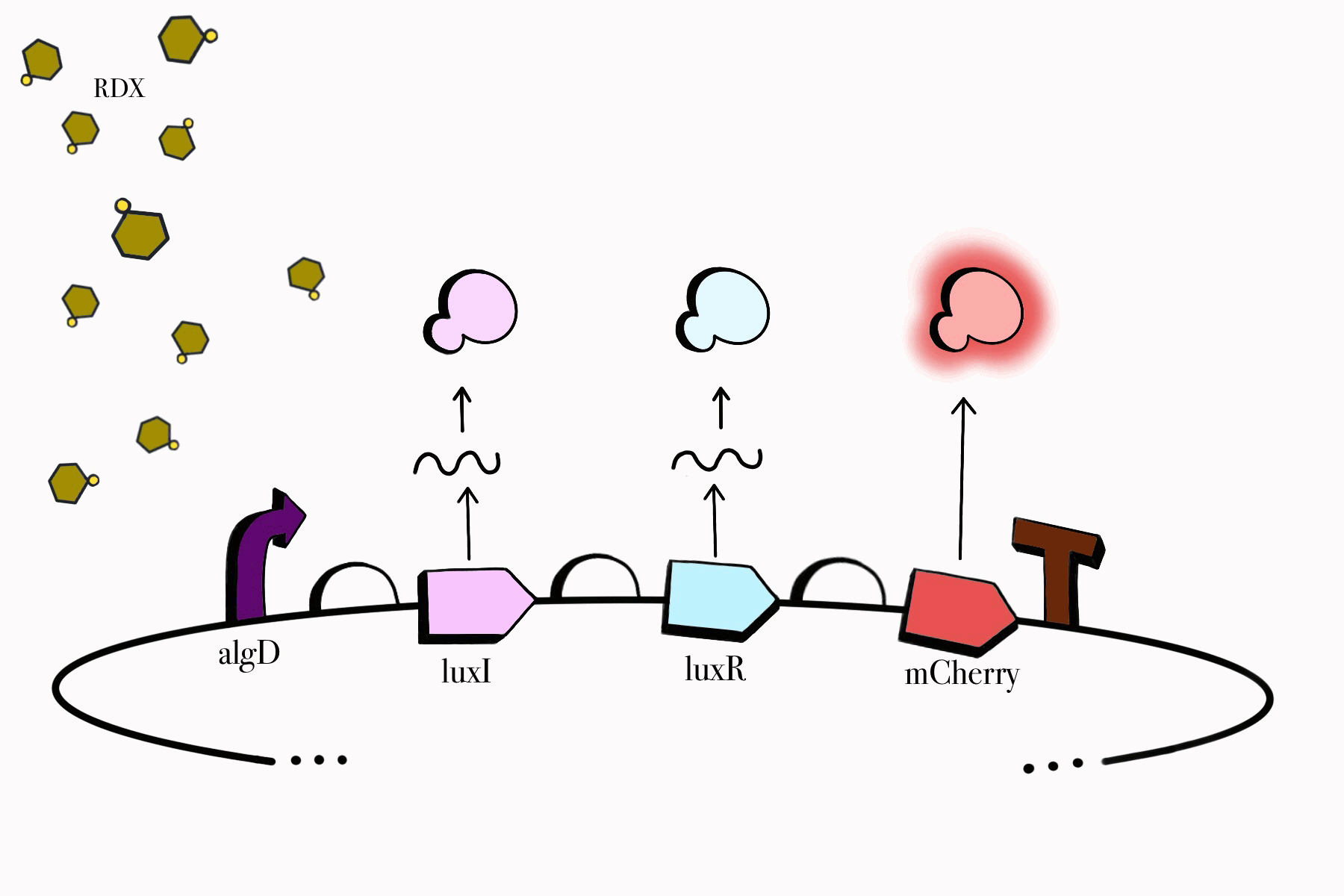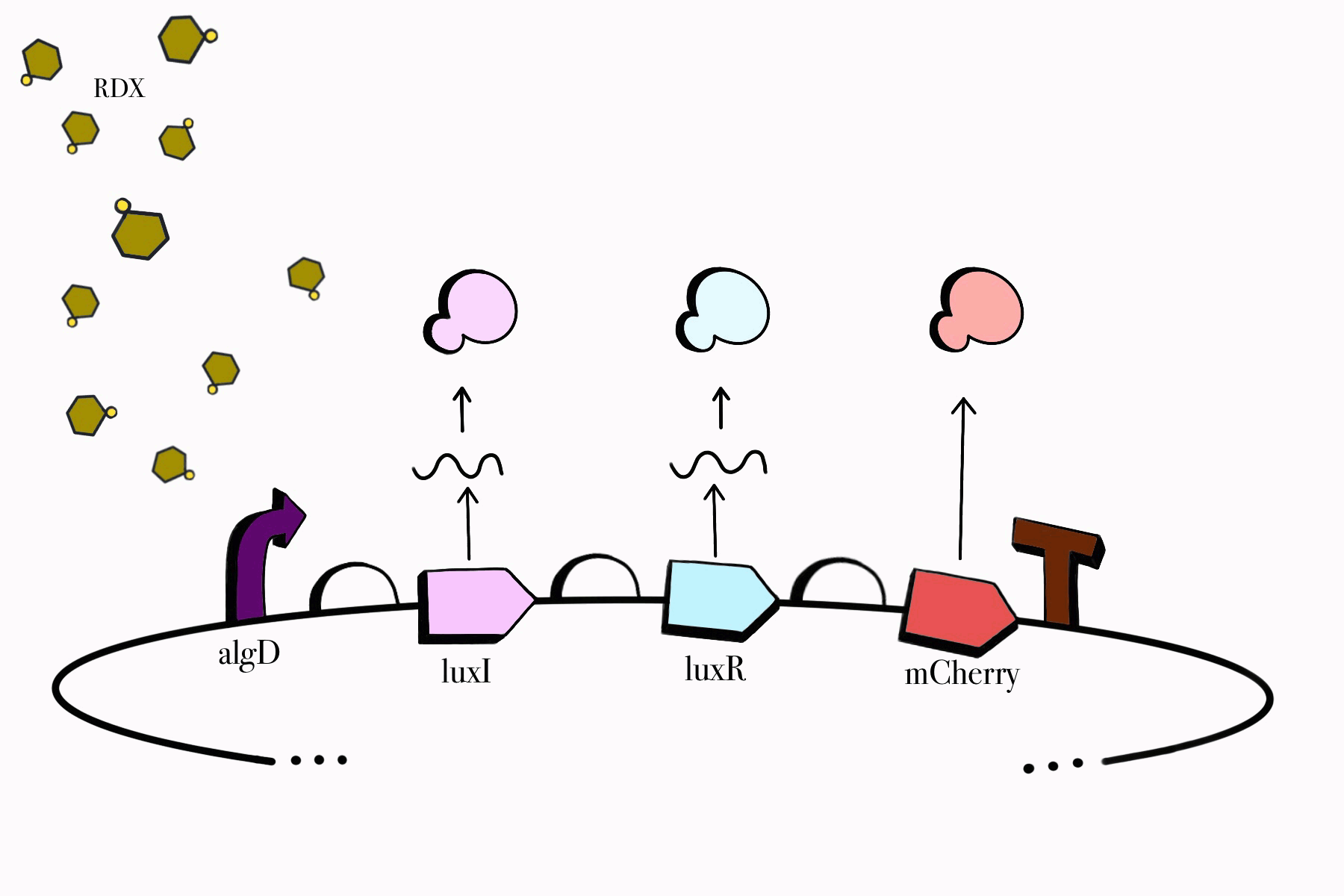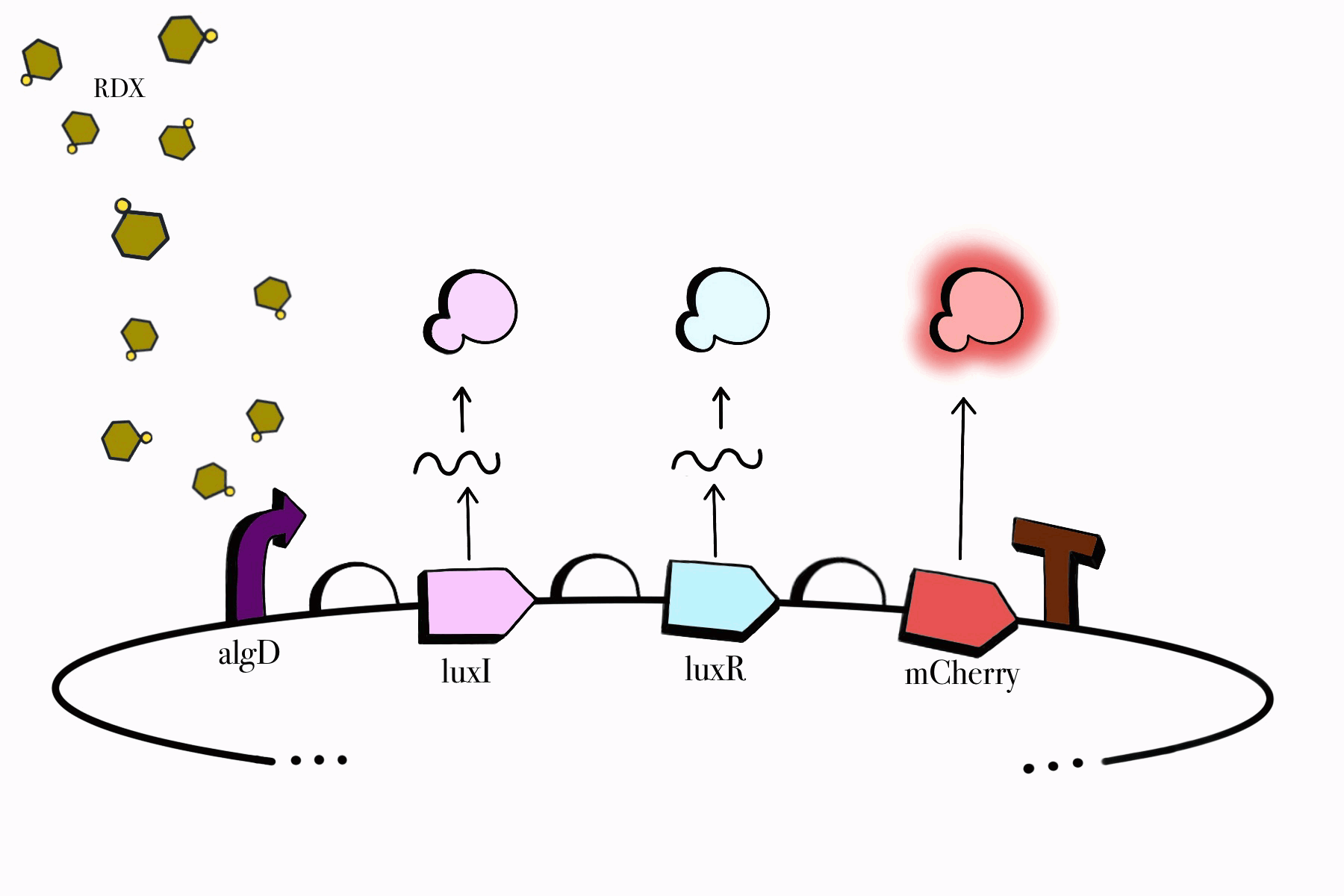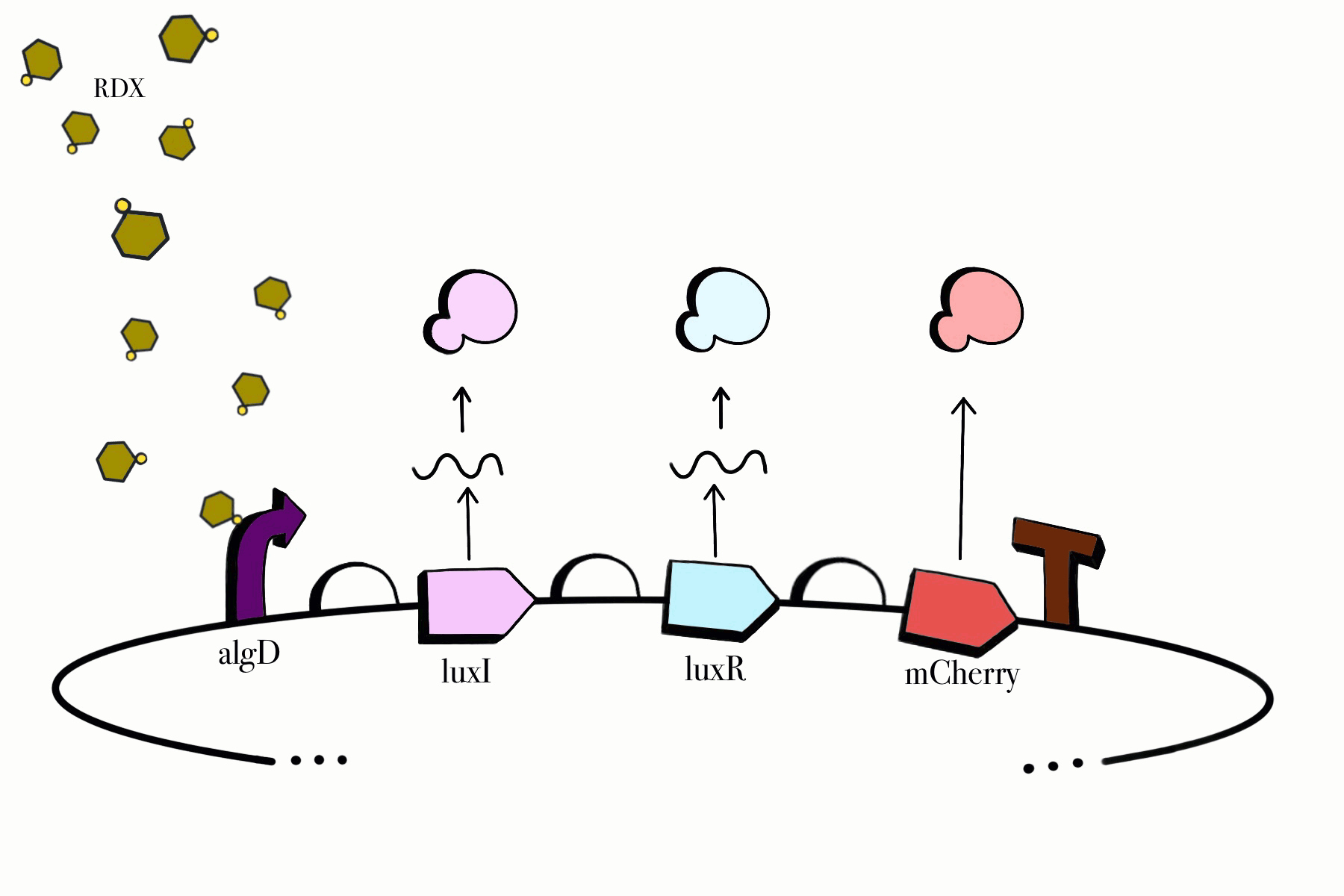
For the first device, the promoter algD is induced by a certain range of concentration (between 0.2-0.5mmolar) of RDX. The sequence of the algD promoter and its regulators were derived from analogous sequences from the genome of the Pseudomonas species. For this cycle, we revised and adjusted the original sequence, which was lacking certain regions that promote the regulation of the promoter algD. Following the promoter are the luxI and luxR genes. These genes will enable the chassis to partake in a quorum sensing system with the second device, which will enable them to act in accordance with the RDX concentrations in their environment. The product of luxI will convert S-adenosylmethionine (SAM) into acyl-homoserine lactone (AHL). Consequently, luxR will produce a protein that binds to AHL. This merge will stimulate the transcription of luxpr (pLuxR) promoter in the second device. To visualize the transcription of this device, the reporter gene mCherry was included, which produces a red fluorescence indicating that gene transcription is operating. Lastly, this sequence ends with the BioBrick terminator.
Device #2: Biodegradation of RDX

Activation of our second device for Biodegradation will be governed by the presences of LuxR bound to AHL, which will act as the inducer for this endeavor. As such, prlux will act as the promoter for this device. When activated, gene expression of xplB and xpla will take place. The xplB gene encodes for a partner flavodoxin reductase, while the xplA encodes for flavodoxin domain fused (at the N-terminus) of a P450 cytochrome. The product of these genes will create a system in charge of the denitrification of RDX taking two possible routes: anaerobic or aerobic pathways, followed by subsequent ring cleavage of RDX. This ring cleavage is essential to produce formaldehyde and nitrite, our secondary products essential for the activation of our third device. When biodegradation of RDX is complete, the gene amilGFP, a yellow fluorescent protein, will function as a reporter gene.
Device #3: Killswitch of RDX Circuit

The third device, considered as a killswitch, is a modified AND gate that is activated by the presence of formaldehyde and nitrite. The presence of formaldehyde activates the pfirm promoter. The pfirm promoter activation then allows the transcription of the supD gene, while the presence of nitrite activates the inducible promoter pyeaR, which allows the transcription of the t7ptag gene. Each promoter possesses a level of regulation due to the nsrr binding sequence. NsrR protein will bind to this specific sequence to repress transcription. Such a sequence is native to the pyeaR. However, due to basal expression detected by NEU_China 2019, an extra binding sequence of nsrr was added after the promoter. In addition, we decided to add this sequence downstream of pfirm to add more regulation to the killswitch. The t7ptag gene codes for a polymerase with two amber or nonsense mutations which inhibits its translation. To modify the amber mutations, the supD gene produces a tRNA that suppresses them. This then activates the P17 promoter and allows expression of the colicin gene. Colicin will cause bacterial lysis, allowing the detection and biodegradation of RDX to cease, and the bacteria to die, promoting biosafety.
References:
Belda, E., van Heck, R. G. A., José Lopez-Sanchez, M., Cruveiller, S., Barbe, V., Fraser, C., Klenk, H.-P., Petersen, J., Morgat, A., Nikel, P. I., Vallenet, D., Rouy, Z., Sekowska, A., Martins dos Santos, V. A. P., de Lorenzo, V., Danchin, A., & Médigue, C. (2016). The revisited genome of Pseudomonas putidaKT2440 enlightens its value as a robust metabolic chassis. Environmental Microbiology, 18(10), 3403–3424. https://doi.org/10.1111/1462-2920.13230
Callahan, S. M., & Dunlap, P. V. (2000). LuxR- and Acyl-Homoserine-Lactone-Controlled Non- lux Genes Define a Quorum-Sensing Regulon in Vibrio fischeri. Journal of Bacteriology, 182(10), 2811–2822. https://doi.org/10.1128/jb.182.10.2811-2822.2000
Chong CS, Sabir DK, Lorenz A, Bontemps C, Andeer P, Stahl DA, Strand SE, Rylott EL, Bruce NC. Analysis of the xplAB-containing gene cluster involved in the bacterial degradation of the explosive hexahydro-1,3,5-trinitro-1,3,5-triazine.Appl Environ Microbiol. 2014 Nov;80(21):6601-10. doi: 10.1128/AEM.01818-14. Epub 2014 Aug 15. PMID: 25128343; PMCID: PMC4249041.
Li, Z., Nair, S.K. (2012). Quorum sensing: how bacteria can coordinate activity and synchronize their response to external signals. Protein science: a publication of the Protein Society, 21(10), 1403-1417. https://doi.org/10.1002/pro.2132
New England Biolabs. (2015). Neb.com. https://international.neb.com/products/c2987-neb-5-alpha-competent-e-coli-high-efficiency#Product%20Information
New England Biolabs. (2021). Neb.com. https://international.neb.com/products/c3019-neb-10-beta-competent-e-coli-high-efficiency#Product%20Information
Sabir DK, Grosjean N, Rylott EL, Bruce NC. Investigating differences in the ability of XplA/B-containing bacteria to degrade explosive hexahedron-1,3,5-trinitro-1,3,5-triazine (RDX). FEMS Micriobiol Lett. 2017 Aug 1;364(14). don: 10.1093/femsle/fnx144. PMID: 28854671.
Images were created with BioRender.com
