Bot for iGEM images uploading
Arriving at the end of their iGEM project, teams accumulated a lot of images, videos, graphs and others. Most of this content is intended to be posted on the wikis. Therefore, people have to enter the igem server and go to the specific page related to document upload. This page is where team members can store their files in iGEM server so that it can be displayed on their wiki, which is hosted by this last server.
The uploading phase is necessary but very repetitive as well. In order to make this process less time-consuming, we had the idea of implementing a python script to do this for you. The package selenium was used for that purpose. The resulting code is able to automatically upload all the files in the iGEM server and save their path into a spreadsheet so that it’s easily accessible. The code has been posted on our GitHub to be available for future iGEM teams. You can check this here.
3D-printed plate for PCR-Echo compatibility
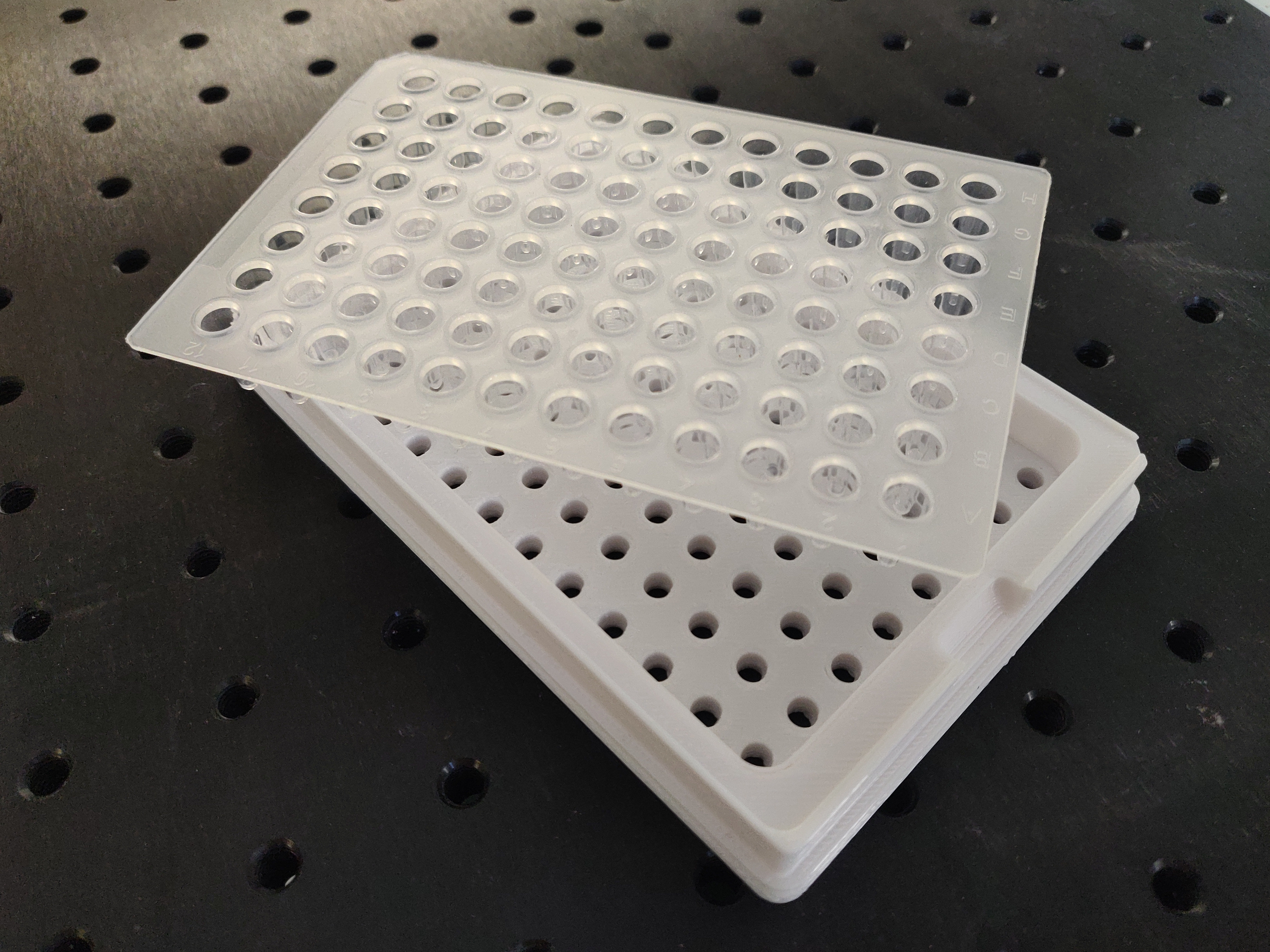
When performing Golden Gate Assembly using the Beckman Coulter machine, we ran into this frankly inconvenient problem: there are no PCR plates available on the market which are compatible with both the Echo machine, and the thermal cyclers available in our lab (BioRad T100 Thermal Cycler). As this specific thermal cycler cannot be used with skirted plates and the Echo machine can only take skirted plates, our first attempts required removing the skirt of the plate using a cutter which is time consuming, potentially harmful and makes the plate unusable for future experiments.
We decided to create a simple 3D printed piece as an adaptor for low-profile non-skirted PCR plates. The adaptor can be easily placed around the PCR plate for it to be compatible with the Echo machine. It can be removed as easily to place the PCR plate in the thermal cycler.
The 3D design files and all required documentation for the printing of the piece are available on GitHub.
You can check this here.
Hardware design
In the context of the current ecological and socio-economical crisis, enabling locally produced chemicals is necessary. Decentralized bio-production of chemicals poses many problems related to bio-safety, sustainability and cost. As minicells lack chromosomal DNA, they are unable to replicate while remaining capable of bio-synthesis. However, working with minicells remains a challenge as it involves many steps, tools and highly trained technicians. To tackle this issue, we propose to automate this process in an affordable bioreactor that does not require the use of chemicals to separate minicells from mother-cells. Here, our implementation of a kill switch strategy based on hardware-synthetic-biology synergies allows for automated triggering of phages-induced cell lysis. To the best of our knowledge, this is the first implementation of a bacteriophage-based strategy for the purification of minicells. By producing this work, we hope to democratize hardware specific to minicells and to enable its usages across disciplines.
Learn more about our hardware design and what it can do in our Hardware page.
Protocols and tips
We used, troubleshooted and optimized many protocols during while creating this project. All of them can be found on our protocols page.
We also created a small QA for performing Golden Gate assembly with an Echo machine. This could be helpful when implementing your own reactions in your lab.
QA for setting up Golden Gate reactions using an Echo machine
When setting up the reaction with the Echo machine, we ran into several problems which we did not account for in the beginning. Some of these issues were particularly difficult to troubleshoot, as there is only a limited amount of literature on them. That is why we think it is even more important to mention these problems here.
Upon reception of plasmids directly in an Echo source plate, transform 1uL to create a glycerol stock of them. This will ensure their availability in case of an incident with the source plate.
The Echo machine can only be used with low-profile skirted PCR plates.
A PCR plate compatible with both the Beckman Coulter Echo™ liquid handler and the thermal cycler available should be used. Plates for the Beckman Coulter Echo™ liquid handler must be skirted and have a standard height whereas the thermal cycler we used could only fit non-skirted plates. To solve this problem, a 3D printed piece was designed to adapt the PCR plate to the Beckman Coulter Echo™ liquid handler.
The PCR plate must be sealed tightly directly after the reactions have been set up. The reaction volumes are around 1uL, low enough for the water to evaporate very quickly. Aluminum seals can be used to prevent evaporation from the plate.
The Cherry Pick software varies depending on the version of the machine. The initial protocol was set up by iGEM Marburg for an Echo 525 but was not compatible with the Echo 550 accessible to our team. The plate type 384_AQ_CP should be favored as it allows calibration for a wide range of fluid types. The protocol should be set up with the following column names: Source plate name, Sample group, Source well, Destination plate name, Destination well, Transfer volume, and Part name (optional). For each liquid transfer, the sample group should be defined. It should be 384PP_Plus_AQ_GP2 for the enzymes (T4 ligase and BsaI-HFv2) and 384PP_Plus_AQ_SP2 for the DNA parts and buffer. The calibration of the machine varies depending on the fluid viscosity. Samples in water require less pulse than samples stored in glycerol which is more viscous. Enzymes are typically stored in 50% glycerol.
A trial using various fluids should be run beforehand to ensure accurate volume transfer by the machine depending on liquid viscosity. A simple protocol can be done using water and 50% glycerol. Various volumes can be transferred to a plate sealed with an aluminum seal. The seal enables a good visualization of the drop transferred and helps determine accurate pipetting. For each fluid type, various volumes can be transferred to different wells and compared.
We unfortunately had to realize that there is no standardized M9 media recipe available for the growth of E.coli. The 2 teams had different protocols. Furthermore, some of the recipes available do not account for the problem of precipitation of the added calcium chloride. If this compound is added too quickly and to low amounts of liquid it will precipitate and therefore be inaccessible to the cell. Therefore, we added calcium chloride as the last ingredient and while agitating using a magnetic stir bar. After extensive comparison of different M9 recipes we decided on one media recipe in order to keep the data more comparable between the two labs.
When conducting the actual measurement using a 384 well plate, we ran into the problem of calibrating the gain for the fluorescence signal of the plate reader. We tried to let the plate reader determine the best gain for the measurement by directing it to a well supplemented with 10µM solution of fluorescein. Unfortunately, the plate reader software encountered a bug, which resulted in a well being chosen as a reference with a very low fluorescence signal. As a consequence, the growth curve resulted in the saturation of fluorescence signal for most wells, making the data unusable.