
Loading...
Choice of Biomarkers
The part of our protocol that we focussed on is

To establish the identity of the biomarkers we can target as well as for establishing thresholds of the biomarkers we’re targeting , we initially established a protocol before moving to a more rigorous data-driven approach.
To better understand the hits that had come from the above study we performed a GO-term analysis on the proteins we obtained from PGI. The genes were sorted by GO-term functionality.
The results are as follows:

There are two categories of proteins that stand out in our query gene set - the “endopeptidase activity” and “peptidase activity” - making up close to ~20% of the genes in the query set. The red bars correspond to the distribution of all genes found in Homo sapiens.
Knowing where to focus, we next chose to explore the genes with the GO-term ‘Molecular Function’.

By sorting with the parameter Molecular Function, we compress the data into broader categories. Although ‘catalytic activity had the second-highest distribution of genes, we chose to go ahead with it as we knew ‘’catalytic activity’’ could be used to make far more robust kits than ‘binding’. We next chose to explore it’.

There are 43 genes under ‘catalytic activity; which show that - i) hydrolase activity is the top choice.

Among the 23 genes under ‘Hydrolase Activity’, ‘’peptidase activity’’ stood out.
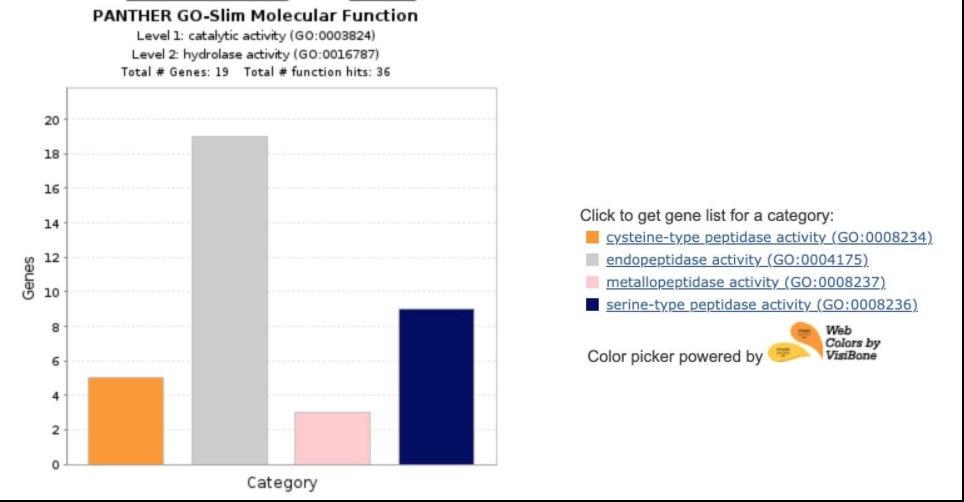
Thus, we concluded that a downstream target of a peptidase could be a potential biomarker.
Next, we queried the list of proteins we had obtained from PGI data from a healthy vs OSCC patient cohort on TCGA to see the quantitative dysregulation of these proteins. This was to get a global picture of the proteins and quantify them according to a globally recognized repository. You can see the list of proteins along with their relative quantification from TCGA,
The top hits from the relative quantification of TCGA RNA-Seq data is shown.
We thus concluded that MMP9 (Matrix Metalloprotease) seemed to be a promising candidate.
It satisfied the three criteria we had set out -
- Listed in the LC-MS data obtained from PGI, Chandigarh
- Compatible with our GO-term enrichment analysis
- Significantly higher expression in tumour saliva samples as against healthy sample
However, to be doubly sure before we made our choice we did another round of literature survey, we found that nearly all publications that had established a link between OSCC and MMP9, had implicated an upregulation in its saliva and/or serum levels in OSCC patients.
On looking at the literature we had surveyed earlier, many had implicated MMP9. Further, it was said to be involved in tumour cell proliferation by clearing the ECM providing more support for its involvement in OSCC.
Thus, we chose MMP9 as our first biomarker in order to establish a proof-of-concept.
