Minghan1005 (Talk | contribs) |
Karin100699 (Talk | contribs) |
||
| (26 intermediate revisions by 4 users not shown) | |||
| Line 22: | Line 22: | ||
</li> | </li> | ||
<li class="nav-item"> | <li class="nav-item"> | ||
| − | <a class="nav-link" href="#Subtitle2"> | + | <a class="nav-link" href="#Subtitle2">Results</a> |
</li> | </li> | ||
</ul> | </ul> | ||
| Line 36: | Line 36: | ||
<!--section name:Overview--> | <!--section name:Overview--> | ||
<h2 id ="Subtitle1">Overview</h2> | <h2 id ="Subtitle1">Overview</h2> | ||
| − | <p> | + | <p>  In our project, the superfolder green fluorescent protein (sfGFP) allows better quantification of promoter strength and sensitivity<sup>[<a href="#ref1">1</a>,<a href="#ref2">2</a>]</sup>. In the oxidative stress sensing system biobrick, we improved the <a href="http://parts.igem.org/Part:BBa_K2610031">biobrick BBa_K2610031</a> from the <a href="https://2019.igem.org/Team:Leiden">2019 iGEM Leiden team</a> by changing GFP (<a href="http://parts.igem.org/Part:BBa_E0040">BBa_E0040</a>)<sup>[<a href="#ref2">2</a>]</sup> into sfGFP(<a href="http://parts.igem.org/Part:BBa_I746916">BBa_I746916</a>), which is hypothesized to have a higher expression level than GFP [Fig. 1]. We also added transcription activator SoxR to our biobrick for increased function of the oxidative stress sensing system<sup>[<a href="#ref3">3</a>]</sup>. |
| − | + | <div class="container-fluid p-0"> | |
| − | + | ||
| − | + | ||
| − | + | ||
<div class="row no-gutters"> | <div class="row no-gutters"> | ||
<div class="col-lg "> | <div class="col-lg "> | ||
<figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
<!-- put link in href & src , width in a can adjust--> | <!-- put link in href & src , width in a can adjust--> | ||
| − | <a href="https://static.igem.org/mediawiki/2021/a/a0/T--NCKU_Tainan--improvement.png" target="_blank" style="width: | + | <a href="https://static.igem.org/mediawiki/2021/a/a0/T--NCKU_Tainan--improvement.png" target="_blank" style="width:70%"><img src="https://static.igem.org/mediawiki/2021/a/a0/T--NCKU_Tainan--improvement.png" alt="" title="" style="width:100%"></a> |
| − | <figcaption>Fig. 1. | + | <figcaption>Fig.1. Improvement big picture</figcaption> |
</figure> | </figure> | ||
</div> | </div> | ||
| Line 55: | Line 52: | ||
<section> | <section> | ||
<!--section name:Experimental Approach & Results--> | <!--section name:Experimental Approach & Results--> | ||
| − | <h2 id ="Subtitle2"> | + | <h2 id ="Subtitle2">Results</h2> |
| − | + | <h3>Disk assay</h3> | |
| − | + | <p>  Disk assay was used to check the effect of each inducer. The concentration of the different inducers we used are listed below:</p> | |
| + | <div class="container-fluid p-0"> | ||
<div class="row no-gutters"> | <div class="row no-gutters"> | ||
<div class="col-lg "> | <div class="col-lg "> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<div class="table-responsive" style="overflow-x:auto;"> | <div class="table-responsive" style="overflow-x:auto;"> | ||
| − | <table class="table-bordered table-striped" style="word-wrap:break-word;"> | + | <table class="table-bordered table-striped" style="word-wrap:break-word; width:50%"> |
<thead> | <thead> | ||
<tr> | <tr> | ||
| − | <th style="text-align: center"> | + | <th style="text-align: center">Inducer</th> |
| − | <th style="text-align: center"> | + | <th style="text-align: center">Volume per disk</th> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</tr> | </tr> | ||
</thead> | </thead> | ||
<tbody> | <tbody> | ||
<tr> | <tr> | ||
| − | + | <td style="text-align: center">H<sub>2</sub>O<sub>2</sub> (30%)</td> | |
| − | + | ||
| − | + | <td style="text-align: center">10 μl</td> | |
| − | + | ||
| − | + | ||
| − | <td style="text-align: center"> | + | |
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="text-align: center"> | + | <td style="text-align: center">H<sub>2</sub>O<sub>2</sub> (3%)</td> |
| − | + | <td style="text-align: center">10 μl</td> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | <td style="text-align: center"> | + | |
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="text-align: center"> | + | <td style="text-align: center">MD (menadione)(10 mM)</td> |
| − | + | <td style="text-align: center">10 μl</td> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="text-align: center"> | + | <td style="text-align: center">DMSO (solvent for MD)</td> |
| − | + | <td style="text-align: center">10 μl</td> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="text-align: center"> | + | <td style="text-align: center">PQ (paraquat)(1 mM)</td> |
| − | + | <td style="text-align: center">10 μl</td> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td style="text-align: center"> | + | <td style="text-align: center">MQ (solvent for PQ)</td> |
| − | + | <td style="text-align: center">10 μl</td> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</tr> | </tr> | ||
| + | |||
</tbody> | </tbody> | ||
</table> | </table> | ||
</div> | </div> | ||
| − | <figcaption> | + | <figcaption>Table 1. Oxidative stress inducers</figcaption> |
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
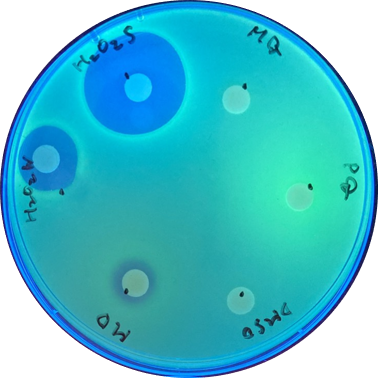
| − | <p> | + | <p>  With sfGFP, the induced system result can be checked under UV light, making it easier to tell the difference between each inducer (Fig. 2).</p> |
| − | + | ||
| + | <div class="container-fluid p-0"> | ||
<div class="row no-gutters"> | <div class="row no-gutters"> | ||
<div class="col-lg "> | <div class="col-lg "> | ||
<figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
<!-- put link in href & src , width in a can adjust--> | <!-- put link in href & src , width in a can adjust--> | ||
| − | <a href="https://static.igem.org/mediawiki/ | + | <a href="https://static.igem.org/mediawiki/2021/8/88/T--NCKU_Tainan--PsoxS_Disk_Assay.png" target="_blank" style="width:50%"><img src="https://static.igem.org/mediawiki/2021/8/88/T--NCKU_Tainan--PsoxS_Disk_Assay.png" alt="" title="" style="width:100%"></a> |
| − | <figcaption>Fig. | + | <figcaption>Fig. 2. The induced result is clear to be seen after the improvement.</figcaption> |
</figure> | </figure> | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <h3>Oxidative Stress Assay</h3> | |
<div class="container-fluid p-0"> | <div class="container-fluid p-0"> | ||
<div class="row no-gutters"> | <div class="row no-gutters"> | ||
| Line 159: | Line 121: | ||
<figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
<!-- put link in href & src , width in a can adjust--> | <!-- put link in href & src , width in a can adjust--> | ||
| − | <a href="https://static.igem.org/mediawiki/ | + | <a href="https://static.igem.org/mediawiki/2021/0/0b/T--NCKU_Tainan--improvement2.png" target="_blank" style="width:70%"><img src="https://static.igem.org/mediawiki/2021/0/0b/T--NCKU_Tainan--improvement2.png" alt="" title="" style="width:100%"></a> |
| − | + | <figcaption>Fig. 3. Biobrick of <i>soxR-P<sub>soxS</sub>-sfgfp</i></figcaption> | |
</figure> | </figure> | ||
</div> | </div> | ||
| Line 166: | Line 128: | ||
</div> | </div> | ||
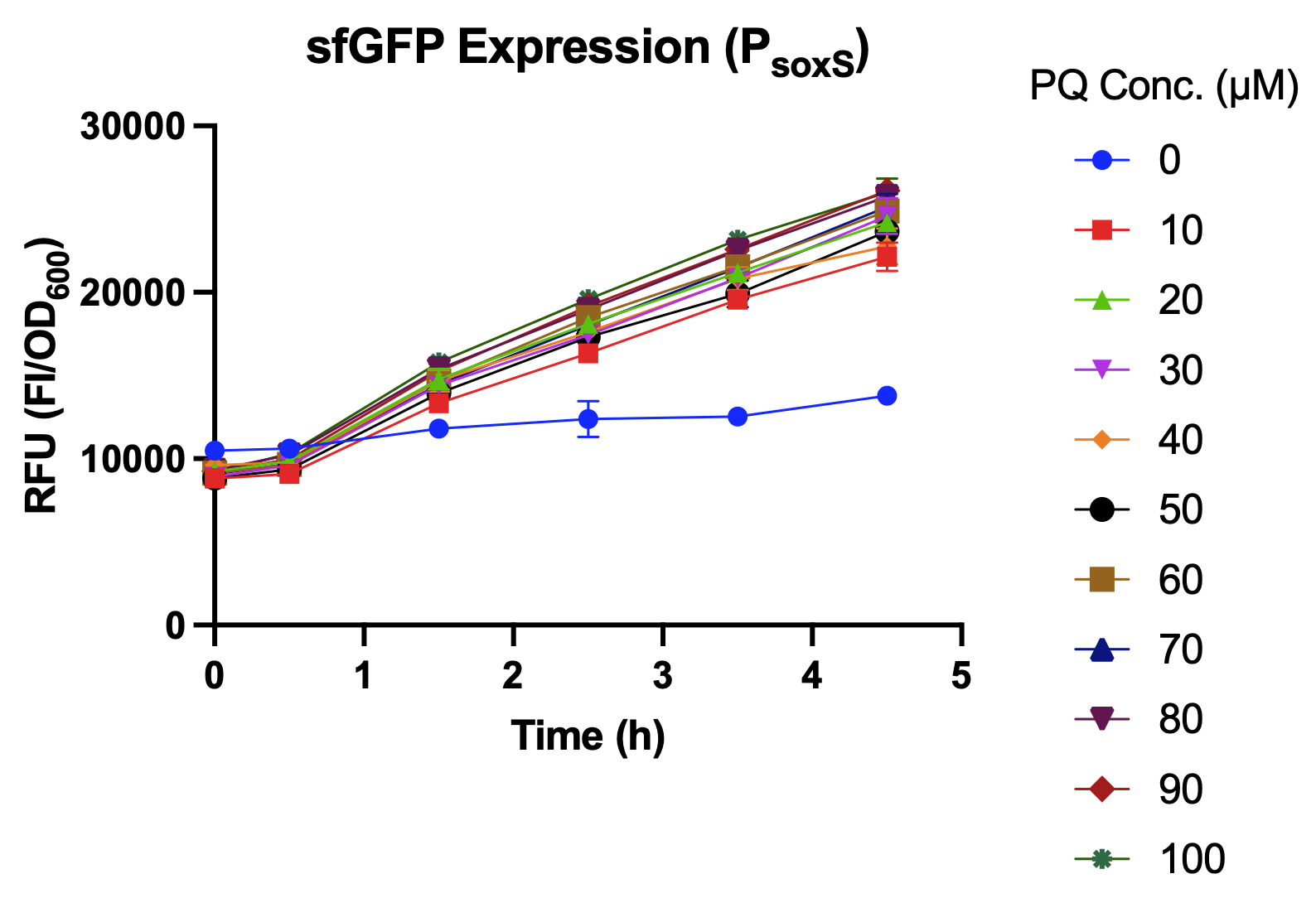
| − | <p> | + | <p>  In this experiment, an overnight culture was prepared, diluted 10 times and transferred into the 96 well plate. The inducer (paraquat-PQ) was added after incubating in 37 °C. Using an ELISA reader, the sfGFP expression level was measured hourly during the 4.5 hours after paraquat induction.</p> |
| − | <p> | + | <p>  We compared sfGFP expression levels of <i>soxS</i> promoter with and without the activation of SoxR transcription factor. As shown in Fig. 3, sfGFP expression levels were higher with SoxR than without SoxR. In addition, there was no leakage problem after we added SoxR into our biobrick<sup>[<a href="#ref4">4</a>]</sup> (Fig. 4).</p> |
| − | + | <div class="container-fluid p-0"> | |
| + | <div class="row no-gutters"> | ||
| + | <div class="col-lg "> | ||
| + | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
| + | <!-- put link in href & src , width in a can adjust--> | ||
| + | <table> | ||
| + | <tr> | ||
| + | <td bgcolor="#FFFAF4"> <a href="https://static.igem.org/mediawiki/parts/6/63/T--NCKU_Tainan--Low_sfGFP_Expression_%28PsoxS%29.png" target="_blank" style="width:50%"><img src="https://static.igem.org/mediawiki/parts/6/63/T--NCKU_Tainan--Low_sfGFP_Expression_%28PsoxS%29.png" alt="" title="" style="width:100%"></a></td> | ||
| + | <td bgcolor="#FFFAF4"><a href="https://static.igem.org/mediawiki/parts/3/33/T--NCKU_Tainan--Del_Low_sfGFP_Expression_%28soxR-PsoxS%29.png" target="_blank" style="width:50%"><img src="https://static.igem.org/mediawiki/parts/3/33/T--NCKU_Tainan--Del_Low_sfGFP_Expression_%28soxR-PsoxS%29.png" alt="" title="" style="width:100%"></a></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <figcaption>Fig. 4. Leakage problem was perfectly solved after the improvement.</figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <p>  Please visit <a href="https://2021.igem.org/Team:NCKU_Tainan/Results">Results page</a> for more information.</p> | ||
| + | |||
| + | |||
| + | </section> | ||
<section class="ref"> | <section class="ref"> | ||
| Line 174: | Line 155: | ||
<ol> | <ol> | ||
<!--id name must corespond --> | <!--id name must corespond --> | ||
| − | <li id="ref1">https:// | + | <li id="ref1">Overkamp W, Beilharz K, Detert Oude Weme R, et al. Benchmarking Various Green Fluorescent Protein Variants in Bacillus subtilis, Streptococcus pneumoniae, and Lactococcus lactis for Live Cell Imaging. <i>Applied and Environmental Microbiology</i>. 2013;79(20):6481-6490. doi:10.1128/aem.02033-13</li> |
| − | <li id=" | + | <li id="ref2"><i>Creation, Expression, Purification and Characterization of GFP G4b GFP Expression and Melting Curves.<a href="https://www.biophysik.physik.uni-muenchen.de/teaching/laboratory_courses/gfp_expression/g4b_gfp_expressionenglish_2017.pdf">https://www.biophysik.physik.uni-muenchen.de/teaching/laboratory_courses/gfp_expression/g4b_gfp_expressionenglish_2017.pdf</a> </i></li> |
| − | + | <li id="ref3">Baez A, Shiloach J. Escherichia coli avoids high dissolved oxygen stress by activation of SoxRS and manganese-superoxide dismutase. <i>Microbial Cell Factories</i>. 2013;12(1):23. doi:10.1186/1475-2859-12-23</li> | |
| − | + | <li id="ref4">Seo S, Kim D, Szubin R, Palsson Bernhard O. Genome-wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks under Oxidative Stress in Escherichia coli K-12 MG1655. <i>Cell Reports</i>. 2015;12(8):1289-1299. doi:10.1016/j.celrep.2015.07.043</li> | |
| + | <li id="ref5">Part:BBa E0040 - parts.igem.org. Igem.org. Published 2013. Accessed October 16, 2021. <a href="http://parts.igem.org/Part:BBa_E0040">http://parts.igem.org/Part:BBa_E0040</a></li> | ||
| + | </ol> | ||
</section> | </section> | ||
</main> | </main> | ||
Latest revision as of 03:50, 14 December 2021

Overview
In our project, the superfolder green fluorescent protein (sfGFP) allows better quantification of promoter strength and sensitivity[1,2]. In the oxidative stress sensing system biobrick, we improved the biobrick BBa_K2610031 from the 2019 iGEM Leiden team by changing GFP (BBa_E0040)[2] into sfGFP(BBa_I746916), which is hypothesized to have a higher expression level than GFP [Fig. 1]. We also added transcription activator SoxR to our biobrick for increased function of the oxidative stress sensing system[3].
Results
Disk assay
Disk assay was used to check the effect of each inducer. The concentration of the different inducers we used are listed below:
| Inducer | Volume per disk |
|---|---|
| H2O2 (30%) | 10 μl |
| H2O2 (3%) | 10 μl |
| MD (menadione)(10 mM) | 10 μl |
| DMSO (solvent for MD) | 10 μl |
| PQ (paraquat)(1 mM) | 10 μl |
| MQ (solvent for PQ) | 10 μl |
With sfGFP, the induced system result can be checked under UV light, making it easier to tell the difference between each inducer (Fig. 2).
Oxidative Stress Assay
In this experiment, an overnight culture was prepared, diluted 10 times and transferred into the 96 well plate. The inducer (paraquat-PQ) was added after incubating in 37 °C. Using an ELISA reader, the sfGFP expression level was measured hourly during the 4.5 hours after paraquat induction.
We compared sfGFP expression levels of soxS promoter with and without the activation of SoxR transcription factor. As shown in Fig. 3, sfGFP expression levels were higher with SoxR than without SoxR. In addition, there was no leakage problem after we added SoxR into our biobrick[4] (Fig. 4).
Please visit Results page for more information.
References
- Overkamp W, Beilharz K, Detert Oude Weme R, et al. Benchmarking Various Green Fluorescent Protein Variants in Bacillus subtilis, Streptococcus pneumoniae, and Lactococcus lactis for Live Cell Imaging. Applied and Environmental Microbiology. 2013;79(20):6481-6490. doi:10.1128/aem.02033-13
- Creation, Expression, Purification and Characterization of GFP G4b GFP Expression and Melting Curves.https://www.biophysik.physik.uni-muenchen.de/teaching/laboratory_courses/gfp_expression/g4b_gfp_expressionenglish_2017.pdf
- Baez A, Shiloach J. Escherichia coli avoids high dissolved oxygen stress by activation of SoxRS and manganese-superoxide dismutase. Microbial Cell Factories. 2013;12(1):23. doi:10.1186/1475-2859-12-23
- Seo S, Kim D, Szubin R, Palsson Bernhard O. Genome-wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks under Oxidative Stress in Escherichia coli K-12 MG1655. Cell Reports. 2015;12(8):1289-1299. doi:10.1016/j.celrep.2015.07.043
- Part:BBa E0040 - parts.igem.org. Igem.org. Published 2013. Accessed October 16, 2021. http://parts.igem.org/Part:BBa_E0040