Overview
Safety instructions and securities have always been the top priorities when it comes to bioengineered food. Although there is still much misinformation on genetically modified organisms (GMO), this motivates us for a more sustainable and safer laboratory and product implementation. We believe that the advancement of synthetic biology comes from responsible acts.
Laboratory Safety
2021 iGEM team NCKU-Tainan is in full compliance with the safety instruction and security policies both at National Cheng Kung University and the iGEM competition
The team conducted experiments in the laboratories of Prof. Masayuki Hashimoto, Prof. Han-Ching Wang, and Prof. I-Son Ng of NCKU which contain equipment of both Biosafety Level 1 and 2. Before entering the labs, all wet members of the team are credited with a “Basic Laboratories Safety Instruction" certificate from National Cheng Kung University after six hours of lectures and an online examination. We also received two weeks of intensive training on equipment and chemical usage, following protocols written by advisors. A supervisor or an instructor was present at all times in the labs.
Project Safety
Escherichia coli Nissle 1917
In order to make our biotherapeutic product safe for human consumption, we used E. coli Nissle 1917 as our chassis, a nonpathogenic E. coli strain isolated by Alfred Nissle in 1917. It is one of the best examined probiotic strains used in many gastrointestinal disorders including diarrhea, uncomplicated diverticular disease, and ulcerative colitis (UC) [1]. It has been proven to have an intestinal anti-inflammatory effect without major immunotoxic properties[2,3]. A clinical study that evaluated the clearance of E. coli Nissle has proven that it does not colonize in the gut for an extended amount of time, making it suitable for probiotic use[4]. Research has shown that daily treatment with E. coli Nissle for chronic disorders is effective.
Environmental Safety
Antibiotics resistance gene is a useful tool for bacterial selection in experiments. However, it is a problem if engineered bacteria with antibiotics resistance genes leak out from laboratories, where plasmids with antibiotics resistance gene interact with other wild-type bacteria and potentially harm our society. As responsible iGEMers, the biosafety of our product must be carefully considered for sustainable production and community.
Since our product is marketed as functional food, we could not control all sold bubbles wrapped with bacteria under a lab-controlled environment. Therefore, for the safety of our customers and environmental protection, we have chosen a fitting solution – the removal of antibiotics resistance genes in our engineered Escherichia coli Nissle 1917 .
While it is simple to remove antibiotics resistance genes, plasmid stability is crucial for our product’s production and quality control. To address both concerns, we have chosen an E. coli Nissle wild-type plasmid named pMUT1[5] to evaluate its plasmid stability without antibiotic resistance genes.
Plasmid pMUT1
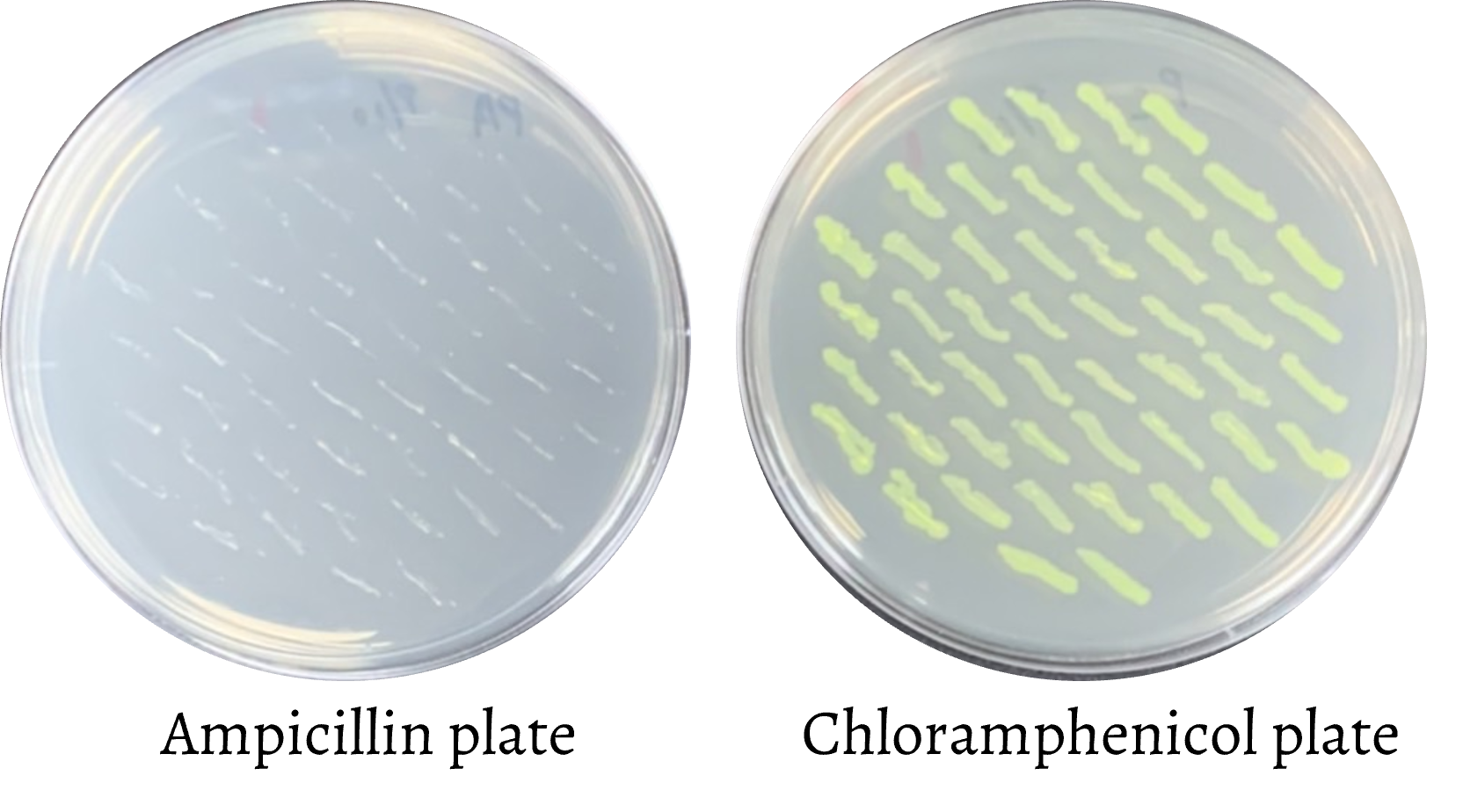
The E. coli Nissle wild-type pMUT1 plasmid does not contain any antibiotic resistance genes. However, for cloning purposes, we added ampicillin resistance gene and superfolder fluorescence protein (sfGFP) gene to the pMUT1 plasmid to select bacterial colonies that contain our desired plasmid. In consideration of biosafety, after successful plasmid construction, the ampicillin resistance gene was removed to ensure that the antibiotic resistance gene does not spread to the outside environment. pMUT1 plasmid stability was also tested to avoid the leakage of the cloned genes into the ecosystem.
The ampicillin resistance gene was removed by using PCR to amplify the whole pMUT1 sequence except for the ampicillin resistance gene. The chloramphenicol resistance gene with two FRT sequences was also amplified by PCR from pKD3 for better selection conditions.
Then, the two fragments were ligated to form a new plasmid with chloramphenicol resistance gene, two FRT sequences, and superfolder fluorescence protein (sfGFP) gene. After electroporation with ligation products, the green colonies on LB agar plate with chloramphenicol were selected and checked by colony PCR.
Soon after, the chloramphenicol resistance gene was also removed by conducting electroporation with pCP20. The connection of pCP20 and FRT sequence was used in this process to delete the gene.
Finally, the pCP20 was easily cured by culturing the bacteria in 42 °C incubator. Only the new plasmid we had designed was kept in the E. coli Nissle.
To further confirm plasmid stability, we conducted a four-passage reinoculation of the bacterial culture, where sfGFP expression and colony numbers are measured every 12 hours for four generations.
| 1st passage | 2nd passage | 3rd passage | 4th passage | 5th passage | |
|---|---|---|---|---|---|
| Sample 1 | 105 Green 9 white |
129 Green 10 white |
174 Green 64 white |
193 Green 121 white |
44 Green 90 white |
| Sample 2 | 105 Green 0 white |
86 Green 0 white |
227 Green 1 white |
302 Green 0 white |
199 Green 1 white |
| Sample 3 | 107 Green 0 white |
115 Green 0 white |
199 Green 0 white |
257 Green 0 white |
145 Green 0 white |
Results
According to the data above, we are confident that pMUT1, an E. coli Nissle wild-type plasmid can retain its plasmid stability with its antibiotic resistance genes removed. From the two successful experimental results, the biosafety of our product is guaranteed for sustainable production.
Three colony samples were chosen to confirm the removal of the antibiotic resistance gene. After five passages, there was a greater number of colonies with no sfGFP expression than those expressing sfGFP in the first sample (Fig. 3), meaning that our desired plasmid was either lost or the bacteria sample was contaminated. There is also a chance that the colonies without sfGFP expression have higher growth rates than those expressing sfGFP. The second and third colonies exhibited sfGFP expression in all five passages (Fig. 4)(Fig. 5). However, since the first sample showed colonies with no sfGFP expression, we cannot confirm the successful removal of the antibiotic resistance gene in the second and third samples as well.
In our future plan to confirm antibiotic resistance gene removal, we plan to choose a positive colony from the first sample and repeat the same experiment. If all colonies express sfGFP, then samples 2 and 3 also hold our desired plasmid, confirming that antibiotic resistance gene removal was successful. However, if negative colonies appear again, this suggests that the plasmids in samples 2 and 3 were most likely lost or the bacteria sample was contaminated.
Device Safety
Escherichia coli Nissle 1917 (EcN)
After finishing each experiment, we will sanitize the waste effluent with bleach to kill all remaining bacteria. Besides, everything that was exposed to E. coli Nissle such as microfluidic tubings and needles will be washed either by MQ water or EtOH and then will be placed inside a biowaste container before throwing away.
Microfluidic Channel
Our microfluidic channel is designed as a disposable product, this is done to avoid any contamination or any bacteria that is still present inside the channel interacting with our test subject when we reuse the microfluidic channel, which will render our experiment result invalid. After the completion of each experiment, we will unplug each tubing connected to the microfluidic channel and immediately sanitize everything with alcohol (EtOH) solutions[6] to kill any remaining bacteria and preventing any possibility of cross-contamination, then all parts will be disposed of in their designated waste container.
References
- Saito, Y., Sato, T., Nomoto, K., & Tsuji, H. (2018). Identification of phenol- and p-Cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiology Ecology, 94(9).
- Zhang, G., Brokx, S., & Weiner, J. H. (2005). Extracellular accumulation of recombinant proteins fused to the carrier protein YebF in Escherichia coli. Nature Biotechnology, 24(1), 100–104.
- Passmore, I. J., Letertre, M., Preston, M. D., Bianconi, I., Harrison, M. A., Nasher, F., … Dawson, L. F. (2018). Para-cresol production by Clostridium difficile affects microbial diversity and membrane integrity of Gram-negative bacteria. PLoS pathogens, 14(9), e1007191.
- Wassenaar TM. Insights from 100 years of research with probioticE. coli. European Journal of Microbiology and Immunology. 2016;6(3):147-161. doi:10.1556/1886.2016.00029
- Lan Y-J, Tan S-I, Cheng S-Y, et al. Development of Escherichia coli Nissle 1917 derivative by CRISPR/Cas9 and application for gamma-aminobutyric acid (GABA) production in antibiotic-free system. Biochemical Engineering Journal. 2021;168:107952. doi:10.1016/j.bej.2021.107952
- Ethanol Tolerance in Bacteria. Critical Reviews in Biotechnology. Published 2021.