Sunnywu0369 (Talk | contribs) |
Pauline 1238 (Talk | contribs) |
||
| (113 intermediate revisions by 10 users not shown) | |||
| Line 11: | Line 11: | ||
<aside class="sidenav d-none d-lg-flex flex-column justify-content-start align-items-center col-2"> | <aside class="sidenav d-none d-lg-flex flex-column justify-content-start align-items-center col-2"> | ||
<ul class="nav flex-column"> | <ul class="nav flex-column"> | ||
| + | <li class="nav-item"> | ||
| + | <a class="nav-link" href="#Subtitle0">Overview</a> | ||
| + | </li> | ||
<li class="nav-item"> | <li class="nav-item"> | ||
<a class="nav-link" href="#Subtitle1">Bubble Production</a> | <a class="nav-link" href="#Subtitle1">Bubble Production</a> | ||
</li> | </li> | ||
<li class="nav-item"> | <li class="nav-item"> | ||
| − | <a class="nav-link" href="#Subtitle2"> | + | <a class="nav-link" href="#Subtitle2">Microfluidic Device</a> |
</li> | </li> | ||
<li class="nav-item"> | <li class="nav-item"> | ||
| − | <a class="nav-link" href="#Subtitle3"> | + | <a class="nav-link" href="#Subtitle3">Sensing for Stress Biomarkers</a> |
</li> | </li> | ||
<li class="nav-item"> | <li class="nav-item"> | ||
| − | <a class="nav-link" href="#Subtitle4"> | + | <a class="nav-link" href="#Subtitle4">Model</a> |
</li> | </li> | ||
</ul> | </ul> | ||
</aside> | </aside> | ||
| + | |||
| + | |||
<main class=""> | <main class=""> | ||
<!--The Page Name--> | <!--The Page Name--> | ||
<br></br> | <br></br> | ||
| − | <img src="https://static.igem.org/mediawiki/2021/ | + | <img src="https://static.igem.org/mediawiki/2021/5/58/T--NCKU_Tainan--pet_header.png" width="87%"> |
<section> | <section> | ||
| − | <p> | + | <p></p> |
| − | + | <h2 id ="Subtitle0">Overview</h2> | |
| + | |||
| + | <p>  The overall goal of MenTAUR is to <b>relieve depressive symptoms by increasing taurine levels in the intestine</b>. To achieve this, we developed multiple models and conducted wet-lab experiments to prove that our bubble, Menbles, can effectively function and release taurine in the human intestine. Menbles are designed to withstand the acidic environment of the stomach, protecting our encapsulated bioengineered <i>E. coli</i>. We developed a microfluidic chip, f(int), that simulated the intestinal environment, allowing us to determine the retention of <i>E. coli</i> in the jejunum. After confirming that most of our <i>E. coli</i> can adhere to the intestinal walls, we proved that the <i>E. coli</i> could detect stress biomarkers reactive oxygen species (ROS) and IFN-γ through the oxidative stress and IFN-γ sensing systems and produce the taurine production enzymes. Finally, with math-based calculations, we can model the mechanism through which taurine converts into TauCl and reduces ROS and IFN-γ levels in the body, relieving depressive symptoms through the gut-brain axis.</p> | ||
| + | |||
| + | |||
| + | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
| + | <!-- put link in href & src , width in a can adjust--> | ||
| + | <a href="https://static.igem.org/mediawiki/2021/6/68/T--NCKU_Tainan--Proof_of_Concept_end.jpg" target="_blank" style="width:90%"><img src="https://static.igem.org/mediawiki/2021/6/68/T--NCKU_Tainan--Proof_of_Concept_end.jpg" alt="" title="" style="width:75%;display:block; margin:auto;" ></a> | ||
| + | <figcaption>Fig. 1. Big picture of Proof of Concept</figcaption> | ||
| + | </figure> | ||
| + | |||
| + | </section> | ||
| + | |||
| + | |||
| + | |||
<section> | <section> | ||
| − | |||
<h2 id ="Subtitle1">Bubble Production</h2> | <h2 id ="Subtitle1">Bubble Production</h2> | ||
| − | + | <p>  Menbles were special edible bubbles that serve as a unique depression medication, coming with a trendy drink worldwide, bubble milk tea. Menbles were designed to be incorporated into everyone’s daily lives, including people who cannot seek help due to stigma. They could quickly get their hands on this drink with functional bubbles.</p> | |
| + | <p>  <b>The biggest challenge to Menbles is the digestion process in the stomach.</b> Menbles must be ensured to withstand the presence of gastric acid in the stomach, as they were designed to release the bacteria in the intestine. Luckily, sodium alginate can withstand these extreme conditions. <b>Sodium alginate has been proven to be resilient even under extreme stomach conditions<sup>[<a href="#ref1">1</a>]</sup>.</b> Therefore, we believe it is ideal for our project to encapsulate our bacteria and not release it until it reaches the intestine, where combined action of acid and trypsin can break the alginate structure down<sup>[<a href="#ref1">1</a>]</sup>, releasing the bacteria.</p> | ||
| + | <p>  However, another challenge we encountered is that <b>although alginate performs very well under acidic conditions, it does not perform too well under basic conditions<sup>[<a href="#ref2">2</a>]</sup>.</b> Therefore, we tested our bubbles containing <i>E. coli</i> Nissle 1917 to be submerged into phosphate buffers of four different pHs, reflecting the range of pHs of milk tea. Furthermore, we tested different bubble sizes because some might experience difficulty swallowing the bubbles without chewing<sup>[<a href="#ref3">3</a>]</sup>. We were then inspired to design the bubbles as tiny as we could. Hence, we conducted experiments using different sizes of bubbles to identify which size has the best recovery rate for our Menbles. Below are the results of the rate of recovery of our bubble for the 14.14 mm<sup>3</sup> (Large) bubble, 4.19 mm<sup>3</sup> (Medium) bubble, and 1.77 mm<sup>3</sup> (Small) bubble (Figure 2). For more detailed information, please refer to the <a href="https://2021.igem.org/Team:NCKU_Tainan/Results" alt="" target="_blank">Results</a> page.</p> | ||
| + | |||
| + | |||
| + | <div class="container-fluid p-0"> | ||
<div class="row no-gutters"> | <div class="row no-gutters"> | ||
| − | <div class="col-lg | + | <div class="col-lg "> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
| − | + | <a href="https://static.igem.org/mediawiki/2021/3/3f/T--NCKU_Tainan--menbleresultcombined.png" target="_blank" style="width:100%"><img src="https://static.igem.org/mediawiki/2021/3/3f/T--NCKU_Tainan--menbleresultcombined.png" alt="" title="" style="width:100%;"></a> | |
| − | <a href="https://static.igem.org/mediawiki/ | + | <figcaption class="mt-3">Fig. 2. The recovery percentage of <i>E. coli</i> Nissle 1917 in bubble with volume 14.14 mm<sup>3</sup> (Large), 4.19 mm<sup>3</sup> (Medium), and 1.77 mm<sup>3</sup> (Small).</figcaption> |
| − | + | ||
| − | <figcaption>Fig. 2. < | + | |
</figure> | </figure> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| − | + | <p>  We hypothesized that as the pH goes higher, the bacteria content in the supernatant is higher due to the breaking down of alginate (in higher pH)<sup>[<a href="#ref2">2</a>]</sup>. Based on our results in Fig. 2, we concluded that the ideal size we would use for Menbles is the 4.19 mm<sup>3</sup> (Medium) bubble because its results best fit our hypothesis. Besides, its bubble recovery is the most stable among other sizes.</p> | |
| − | + | ||
</section> | </section> | ||
| + | <br> | ||
<!--divider--> | <!--divider--> | ||
<hr> | <hr> | ||
<section> | <section> | ||
| − | <h2 id ="Subtitle2"> | + | <h2 id ="Subtitle2">Microfluidic Device</h2> |
<div class="container-fluid p-0" > | <div class="container-fluid p-0" > | ||
<div class="row no-gutters"> | <div class="row no-gutters"> | ||
| − | + | <p>  After we consume the engineered <i>E. coli</i> Nissle bubbles, there are almost no function or model that able to predict the number of probiotics remain in the human body accurately. As a result, <b>we intended to take the retention ratio of <i>E. coli</i> Nissle in the jejunum into consideration</b>. Nevertheless, due to the restriction of human trials and ethical issues, we came up with an idea to invent <b>f(int)</b> - a microfluidic channel to simulate the jejunum environment.</p> | |
| − | + | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | |
| − | + | <!-- put link in href & src , width in a can adjust--> | |
| − | + | <a href="https://static.igem.org/mediawiki/2021/1/14/T--NCKU_Tainan--ProofOfConceptf%28int%29bubbleinside.jpg " target="_blank" style="width:90%"><img src="https://static.igem.org/mediawiki/2021/1/14/T--NCKU_Tainan--ProofOfConceptf%28int%29bubbleinside.jpg " alt="" title="" style="width:80%;display:block; margin:auto;" ></a> | |
| − | + | <figcaption>Fig. 3. f(int) close up view during the experiment</figcaption> | |
| + | </figure> | ||
| + | </div> | ||
| + | <p>  In our Generation 1 experimental results , as shown in Fig. 4, we found that the retention ratio of microfluidic chip w/ CF+2villi is higher than any other channel. In addition, when we compare the result of the same channel with additional 5% HA (Hyaluronic acid) that will be the substitution of mucus, the retention ratio increases by 5.45% (From 80.69% to 86.14%).</p> | ||
| + | </div> | ||
| + | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
| + | <!-- put link in href & src , width in a can adjust--> | ||
| + | <a href="https://static.igem.org/mediawiki/2021/a/a7/T--NCKU_Tainan--G1Ratio.png" target="_blank" style="width:90%"><img src="https://static.igem.org/mediawiki/2021/a/a7/T--NCKU_Tainan--G1Ratio.png" alt="" title="" style="width:80%;display:block; margin:auto;" ></a> | ||
| + | <figcaption>Fig. 4. The retention ratio of different microfluidic channels. (w/ CF (structure contains circular folds); w/ CF+villus means (structure contains villus per circular fold); w/ CF+2villi (structure contains 2villi per circular fold) )</figcaption> | ||
| + | </figure> | ||
| + | |||
| + | <p>  The approach that we took to use 5% HA is because jejunum intestinal juice has 0.2 to 5% mucin(MUC2)<sup>[<a href="#ref4">4</a>]</sup>, and HA was used because of its same sticky characteristic with mucin and due to its low availability. Our experiment result shows that the microfluidic channel with a complex structure such as channel w/ CF+2villi that includes additional HA is the most suitable for simulating the human intestine.</p> | ||
| + | </section> | ||
| + | <!--divider--> | ||
| + | <hr> | ||
| + | <section> | ||
| + | <h2 id ="Subtitle3">Sensing for Stress Biomarkers</h2> | ||
| + | <h3><b>Oxidative Stress Sensing System Experiments</b></h3> | ||
| + | <p>  After confirming that most of the engineered bacteria can adhere to the walls of the intestine, we must prove that the bacteria can produce taurine. We designed our engineered bacteria to produce these enzymes only when the body is under high-stress levels, which are signaled by high levels of reactive oxygen species (ROS) and IFN-γ. <b>The following experiments were performed to confirm that both oxidative stress and IFN-γ sensing systems can effectively detect their respective stress biomarkers and initiate taurine production.</b> </p> | ||
| + | <div class="container-fluid p-0"> | ||
| + | <div class="row no-gutters"> | ||
| + | <div class="col-lg "> | ||
| + | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
| + | <!-- put link in href & src , width in a can adjust--> | ||
| + | <a href="https://static.igem.org/mediawiki/2021/e/ea/T--NCKU_Tainan--oxidative_stress_csad.jpg" target="_blank" style="width:90%"><img src="https://static.igem.org/mediawiki/2021/e/ea/T--NCKU_Tainan--oxidative_stress_csad.jpg" alt="" title="" style="width:100%"></a> | ||
| + | <figcaption>Fig. 5. Mechanism of the oxidative stress sensing system</figcaption> | ||
| + | </figure> | ||
</div> | </div> | ||
| − | + | </div> | |
| + | </div> | ||
| + | <p>  The oxidative stress sensing system regulates the production of L-cysteine sulfinic acid decarboxylase (CSAD). It consists of the <i>soxR</i> gene (<a href="http://parts.igem.org/Part:BBa_K3771100" target="_blank">BBa_K3771100</a>), promoter <i>P<sub>soxS</sub></i> (<a href="http://parts.igem.org/Part:BBa_K3771048" target="_blank">BBa_K3771048</a>), and the <i>csad</i> gene (<a href="http://parts.igem.org/Part:BBa_K3771008" target="_blank">BBa_K3771008</a>). When SoxR protein is fully oxidized, it becomes a powerful transcription activator of <i>P<sub>soxS</sub></i>, leading to the expression of the downstream gene. To test the intensity and specificity of the oxidative stress sensing system, the oxidative stress assay was performed.</p> | ||
| + | <h4><b>Oxidative Stress Assay</b></h4> | ||
| + | <h5><b>Achievements</b></h5> | ||
| + | <p>1. Compared the intensity and sensitivity of different promoters under the stimulation by oxidative stress.</p> | ||
| + | <p>2. Selected a suitable promoter for regulating the expression of L-cysteine sulfinic acid decarboxylase (CSAD).</p> | ||
| + | <h5><b>Process</b></h5> | ||
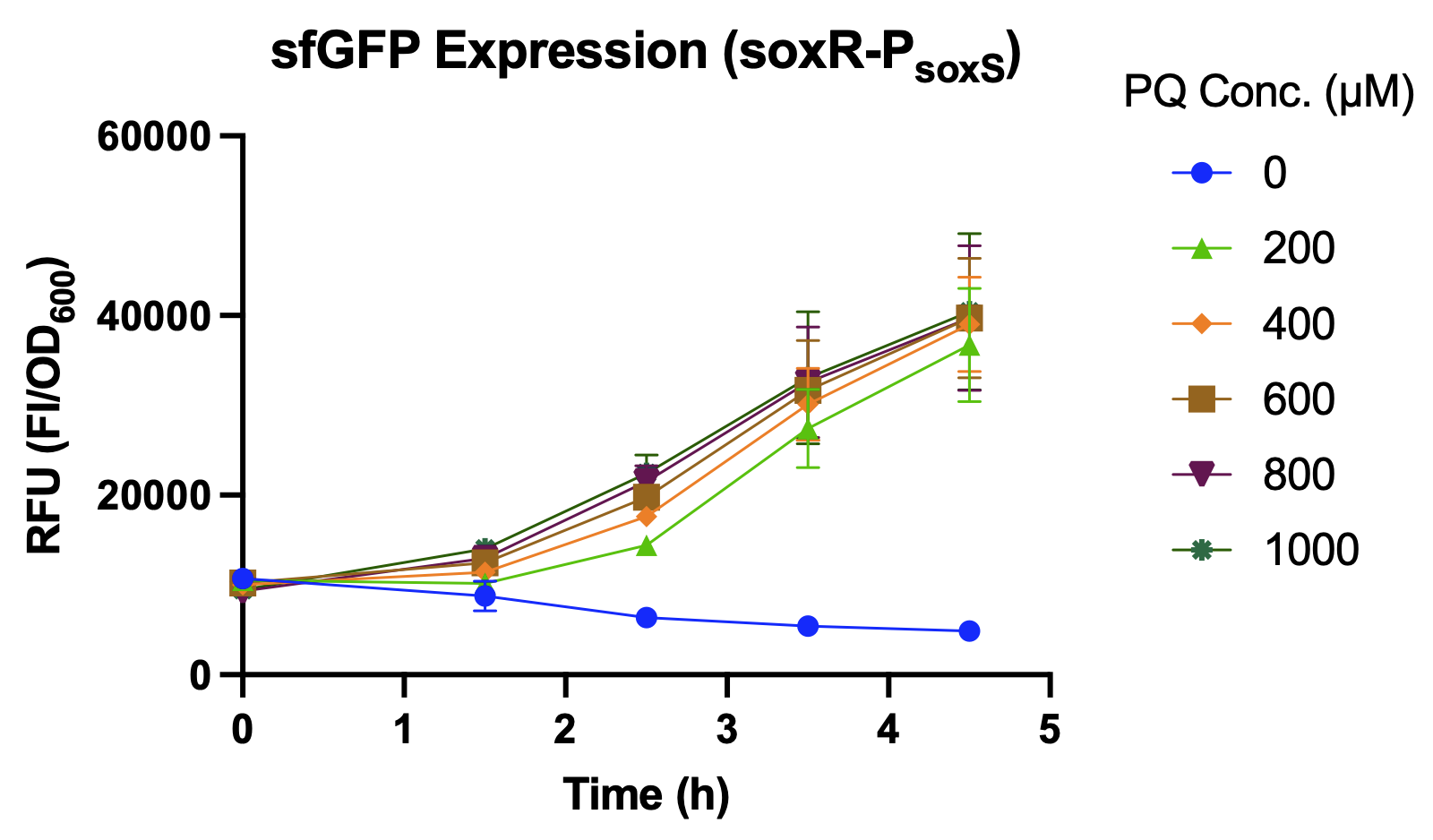
| + | <p>  We compared the relative fluorescence intensity of sfGFP expressed by different promoters when induced by different concentrations of paraquat (PQ). Paraquat can react with oxygen to produce various types of reactive oxygen species (ROS), such as superoxide radical (O<sub>2</sub><sup>−</sup>•), hydrogen peroxide (H<sub>2</sub>O<sub>2</sub>), and hydroxyl radical (OH<sup>−</sup>•)<sup>[<a href="#ref5">5</a>]</sup>, which is used to simulate abnormal oxidative stress in the intestines of stressed individuals. </p> | ||
| + | <p>  Over a period of 4.5 hours after paraquat was added, the relative fluorescence intensity was measured. J23100, a constitutive promoter, being the positive control, shows no difference in sfGFP expression level between the samples with and without paraquat. The activity of <i>P<sub>soxS</sub></i> (<a href="http://parts.igem.org/Part:BBa_K3771048" target="_blank">BBa_K3771048</a>) is higher than that of <i>P<sub>katG</sub></i> (<a href="http://parts.igem.org/Part:BBa_K3771047" target="_blank">BBa_K3771047</a>), but the activity is greatest when the SoxR transcription factor (<a href="http://parts.igem.org/Part:BBa_K3771100" target="_blank">BBa_K3771100</a>) is added in the construction together with <i>P<sub>soxS</sub></i> (Fig. 6).</p> | ||
| + | <div class="container-fluid p-0" > | ||
| + | <div class="row no-gutters"> | ||
| + | <div class="col-lg"> | ||
<figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
| − | + | <a href="https://static.igem.org/mediawiki/parts/c/c2/T--NCKU_Tainan--sfGFP_Expression_%28PQ_%404.5h%29.png" target="_blank" style="width:70%"><img src="https://static.igem.org/mediawiki/parts/c/c2/T--NCKU_Tainan--sfGFP_Expression_%28PQ_%404.5h%29.png" alt="" title="" style="width:100%"></a> | |
| − | <a href="https://static.igem.org/mediawiki/ | + | <figcaption class="mt-3">Fig. 6. sfGFP expression under high concentration of PQ</figcaption> |
| − | <figcaption>Fig. | + | </figure> |
| + | </div> | ||
| + | <div class="col-lg d-flex justify-content-center align-items-center"> | ||
| + | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
| + | <a href="https://static.igem.org/mediawiki/parts/0/0d/T--NCKU_Tainan--Low_sfGFP_Expression_%28PQ_%404.5h%29.png" target="_blank" style="width:70%"><img src="https://static.igem.org/mediawiki/parts/0/0d/T--NCKU_Tainan--Low_sfGFP_Expression_%28PQ_%404.5h%29.png" alt="" title="" style="width:110%"></a> | ||
| + | <figcaption class="mt-3">Fig. 7. sfGFP expression under lower concentration of PQ. The comparison of different promoter strengths by relative fluorescence intensity after 4.5-hour incubation with paraquat in various concentrations was shown in these two pictures.</figcaption> | ||
</figure> | </figure> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | </div> | + | </div> |
| − | + | <p>  Another oxidative stress assay was performed only with SoxR and <i>P<sub>soxS</sub></i> promoter to determine the efficiency of this detection system. The sfGFP level was measured hourly during the 4.5 hours after paraquat induction, and results showed that even low concentrations of paraquat were able to activate the detection system (Fig. 8 and 9).</p> | |
| + | <div class="container-fluid p-0" > | ||
<div class="row no-gutters"> | <div class="row no-gutters"> | ||
| − | <div class="col-lg | + | <div class="col-lg"> |
<figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
| − | + | <a href="https://static.igem.org/mediawiki/parts/3/33/T--NCKU_Tainan--Del_Low_sfGFP_Expression_%28soxR-PsoxS%29.png" target="_blank" style="width:70%"><img src="https://static.igem.org/mediawiki/parts/3/33/T--NCKU_Tainan--Del_Low_sfGFP_Expression_%28soxR-PsoxS%29.png" alt="" title="" style="width:100%"></a> | |
| − | <a href="https://static.igem.org/mediawiki/ | + | <figcaption class="mt-3">Fig. 8. sfGFP expression under low concentration of PQ.</figcaption> |
| − | <figcaption>Fig. | + | |
</figure> | </figure> | ||
</div> | </div> | ||
| − | <div class="col-lg- | + | <div class="col-lg d-flex justify-content-center align-items-center"> |
| − | < | + | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> |
| − | + | <a href="https://static.igem.org/mediawiki/parts/0/0c/T--NCKU_Tainan--Del_High_sfGFP_Expression_%28soxR-PsoxS%29.png" target="_blank" style="width:70%"><img src="https://static.igem.org/mediawiki/parts/0/0c/T--NCKU_Tainan--Del_High_sfGFP_Expression_%28soxR-PsoxS%29.png" alt="" title="" style="width:100%"></a> | |
| − | + | <figcaption class="mt-3">Fig. 9. sfGFP expression under high concentration of PQ. Relative fluorescence intensity of <i>soxR-P<sub>soxS</sub></i> after 4.5-hour incubation with paraquat in various concentrations was shown in these two pictures.</figcaption> | |
| + | </figure> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | </div> | + | </div> |
| − | + | <h3><b>Interferon-gamma Sensing System Experiments</b></h3> | |
| − | + | <p>  The second detection system is the IFN-γ sensing system, which regulates the production of CS. </p> | |
| − | + | <div class="container-fluid p-0"> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<div class="row no-gutters"> | <div class="row no-gutters"> | ||
<div class="col-lg "> | <div class="col-lg "> | ||
<figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
| − | + | <a href="https://static.igem.org/mediawiki/2021/f/f9/T--NCKU_Tainan--IFN-r_system_1234.gif" target="_blank" style="width:100%"><img src="https://static.igem.org/mediawiki/2021/f/f9/T--NCKU_Tainan--IFN-r_system_1234.gif" alt="" title="" style="width:100%"></a> | |
| − | <a href="https://static.igem.org/mediawiki/ | + | <figcaption class="mt-3">Fig. 10. Mechanism of IFN-γ sensing system</figcaption> |
| − | <figcaption>Fig. | + | |
</figure> | </figure> | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <p>  The pathway for the IFN-γ sensing system requires two steps. First, <i>ompA</i> promoter constitutively expresses the OmpA/OprF chimeric protein, which is located in the plasma membrane of <i>E. coli</i>. Then, IFN-γ binds to the chimeric protein, activating a series of signal transduction reactions and finally inducing the <i>pspA</i> promoter and producing the CS enzyme. An IFN-γ induction assay was performed to confirm the function of the IFN-γ sensing system. </p> | |
| − | + | <h4><b>IFN-γ Induction Assay</b></h4> | |
| − | + | <h5><b>Achievements</b></h5> | |
| + | <p>  1. Selected a suitable promoter for regulating the expression of chimeric OmpA/OprF protein</p> | ||
| + | <p>  2. Determined the minimum IFN-γ concentration required for activation of the sensing system</p> | ||
| + | <p>  3. Determined the time required for the system to produce a significant difference in protein expression</p> | ||
| + | <h5><b>Process</b></h5> | ||
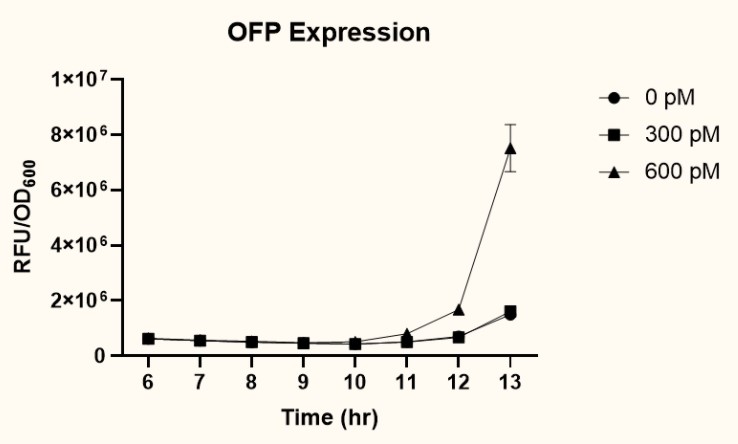
| + | <p>  The two plasmids <i>P<sub>pspA</sub>-ofp</i>-<i>P<sub>lacI</sub>-ompA/oprF</i> (<a href="http://parts.igem.org/Part:BBa_K3771017" target="_blank">BBa_K3771017</a>) and <i>P<sub>pspA</sub>-ofp</i>-<i>P<sub>ompA</sub>-ompA/oprF</i> (<a href="http://parts.igem.org/Part:BBa_K3771018" target="_blank">BBa_K3771018</a>) were transformed into <i>ompA</i> gene knockout strain, JW0940, to prevent <i>OmpA</i> from interfering with IFN-γ binding to chimeric OmpA/OprF. Human IFN-γ of concentrations 300 pM and 600 pM were added to bacteria cultures to induce the system. OFP expression (RFU) was recorded using a microplate reader and normalized by OD<sub>600</sub>. As shown in Fig. 11 and 12, starting at around 12 hours after induction, OFP expression of both samples increased dramatically. </p> | ||
| + | <div class="container-fluid p-0" > | ||
| + | <div class="row no-gutters"> | ||
| + | <div class="col-lg"> | ||
| + | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
| + | <a href="https://static.igem.org/mediawiki/2021/a/ab/T--NCKU_Tainan--proof_of_concept_4.jpg" target="_blank" style="width:70%"><img src="https://static.igem.org/mediawiki/2021/a/ab/T--NCKU_Tainan--proof_of_concept_4.jpg" alt="" title="" style="width:100%"></a> | ||
| + | <figcaption class="mt-3">Fig. 11. OFP expression of <i>P<sub>pspA</sub>-ofp-P<sub>lacI<sub>-ompA/oprF </i>(sample 1) (BBa_K3771017) </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | <div class="col-lg d-flex justify-content-center align-items-center"> | ||
| + | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
| + | <a href="https://static.igem.org/mediawiki/2021/a/a3/T--NCKU_Tainan--proof_of_concept_5.jpg" target="_blank" style="width:70%"><img src="https://static.igem.org/mediawiki/2021/a/a3/T--NCKU_Tainan--proof_of_concept_5.jpg" alt="" title="" style="width:100%"></a> | ||
| + | <figcaption class="mt-3">Fig. 12. OFP expression of <i>P<sub>pspA</sub>-ofp-P<sub>ompA</sub>-ompA/oprF</i> (sample 2) (BBa_K3771018)</figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <p>  As presented in Fig. 13, at 12 hours after induction, sample 2 (<i>P<sub>pspA</sub>-ofp-P<sub>ompA</sub>-ompA/oprF</i>) (<a href="http://parts.igem.org/Part:BBa_K3771018" target="_blank">BBa_K3771018</a>) with the addition of 600 pM IFN-γ had higher OFP expression than that of sample 1 (<i>P<sub>pspA</sub>-ofp</i>-P<i><sub>lacI</sub>-ompA/oprF</i>) (<a href="http://parts.igem.org/Part:BBa_K3771017" target="_blank">BBa_K3771017</a>). Since <i>ompA</i> promoter stimulates higher protein expression than <i>lacI</i> promoter, we decided to incorporate the <i>ompA</i> promoter in our final biobrick to regulate the expression of <i>cs</i>. </p> | ||
| + | <div class="container-fluid p-0"> | ||
| + | <div class="row no-gutters"> | ||
| + | <div class="col-lg "> | ||
| + | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
| + | <a href="https://static.igem.org/mediawiki/2021/1/1a/T--NCKU_Tainan--ifn_12h.jpg" target="_blank" style="width:50%"><img src="https://static.igem.org/mediawiki/2021/1/1a/T--NCKU_Tainan--ifn_12h.jpg" alt="" title="" style="width:100%"></a> | ||
| + | <figcaption class="mt-3">Fig. 13. OFP expression of sample 1 (<i>P<sub>pspA</sub>-ofp-P<sub>lacI</sub>-ompA/oprF</i>) (BBa_K3771018) and sample 2 (<i>P<sub>pspA</sub>-ofp-P<sub>ompA</sub>-ompA/oprF</i>) (BBa_K3771018)</figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | </section> | ||
| + | |||
| + | |||
| + | |||
<section> | <section> | ||
| − | <h2 id ="Subtitle4"> | + | <h2 id ="Subtitle4">Model</h2> |
| − | <p> | + | <p>  Since reducing the amount of ROS was the main goal of Menbles, the mechanism we proposed was the anti-oxidative characteristic of the taurine derivative, TauCl. TauCl could induce lots of antioxidants to be produced within the intestinal epithelial cell. We modeled how GSH, one of the main antioxidants, could relieve ROS (see Fig. 14) in a faster and stronger way. Followed by the physiological mechanism proposed in <a href="https://2021.igem.org/Team:NCKU_Tainan/Description" target="_blank">Description</a> a healthy gut could bi-directionally influence the brain, ultimately reducing various depressive symptoms. </p> |
<div class="container-fluid p-0"> | <div class="container-fluid p-0"> | ||
<div class="row no-gutters"> | <div class="row no-gutters"> | ||
| Line 143: | Line 214: | ||
<figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | <figure class="d-flex flex-column justify-content-center align-items-center px-lg-3"> | ||
<!-- put link in href & src , width in a can adjust--> | <!-- put link in href & src , width in a can adjust--> | ||
| − | <a href="https://static.igem.org/mediawiki/ | + | <a href="https://static.igem.org/mediawiki/2021/7/75/T--NCKU_Tainan--compare.jpg" target="_blank" style="width:80%"><img src="https://static.igem.org/mediawiki/2021/7/75/T--NCKU_Tainan--compare.jpg" alt="" title="" style="width:80%"></a> |
| − | <figcaption>Fig. | + | <figcaption>Fig. 14. The efficiency of excessive ROS scavenging system between endogenous system and enhanced system. Normal intracellular ROS ranges from 2.6-2 μM.</figcaption> |
</figure> | </figure> | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | ||
| − | + | ||
</section> | </section> | ||
| + | <section class="ref"> | ||
| + | <h2>References</h2> | ||
| + | <ol> | ||
| + | <li id="ref1">Guo L, Goff HD, Xu F, et al. The effect of sodium alginate on nutrient digestion and metabolic responses during both in vitro and in vivo digestion process. <i>Food Hydrocolloids</i>. 2020;107:105304. doi:10.1016/j.foodhyd.2019.105304</li> | ||
| + | <li id="ref2">Chuang J-J, Huang Y-Y, Lo S-H, et al. Effects of pH on the Shape of Alginate Particles and Its Release Behavior. <i>International Journal of Polymer Science</i>. 2017;2017:1-9. doi:10.1155/2017/3902704</li> | ||
| + | <li id="ref3">Aldridge KJ, Taylor NF. Dysphagia is a Common and Serious Problem for Adults with Mental Illness: A Systematic Review. <i>Dysphagia</i>. 2011;27(1):124-137. doi:10.1007/s00455-011-9378-5</li> | ||
| + | <li id="ref4">Leal J, Smyth HDC, Ghosh D. Physicochemical properties of mucus and their impact on transmucsal drug delivery. <i>International Journal of Pharmaceutics</i>. 2017;532(1):555-572. doi:10.1016/j.ijpharm.2017.09.018</li> | ||
| + | <li id="ref5">Lascano R, Muñoz N, Robert G, et al. Paraquat: an oxidative stress inducer. <i>Herbicides-properties, synthesis and control of weeds.</i> 2012:135-148. </li> | ||
| + | </ol> | ||
| + | </section> | ||
| + | |||
</main> | </main> | ||
| − | + | <div> | |
| − | + | </div> | |
| − | <script src="https:// | + | |
| − | <script src="https:// | + | <script src="https://2019.igem.org/Team:NCKU_Tainan/js/jquery-3_4_1_min_js?action=raw&ctype=text/javascript"></script> |
| − | <script src="https:// | + | <script src="https://2019.igem.org/Team:NCKU_Tainan/js/4.3.1/bootstrap_min_js?action=raw&ctype=text/javascript"></script> |
| + | <script src="https://2019.igem.org/Team:NCKU_Tainan/js/Universal?action=raw&ctype=text/javascript"></script> | ||
</body> | </body> | ||
Latest revision as of 07:18, 8 December 2021

Overview
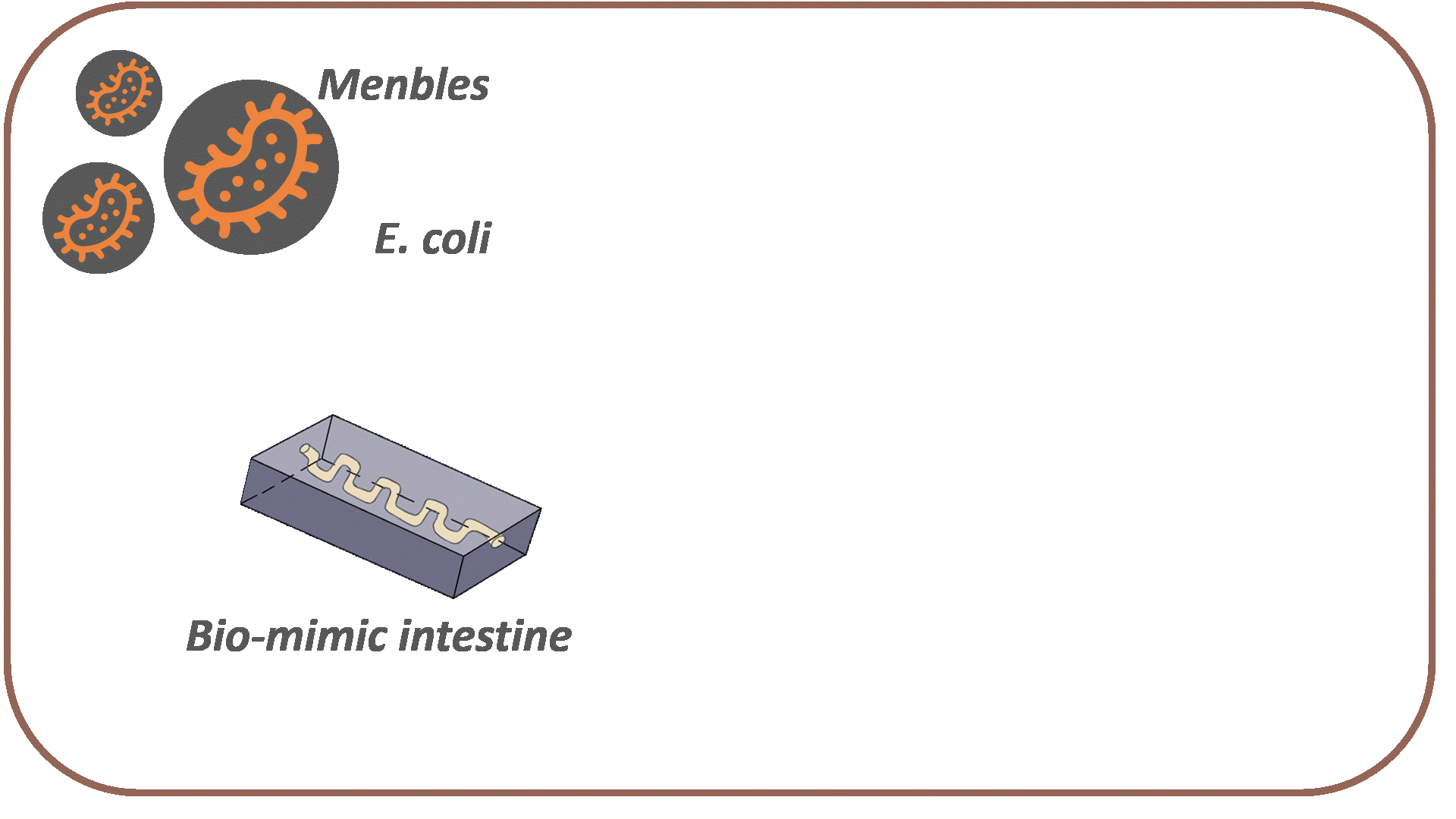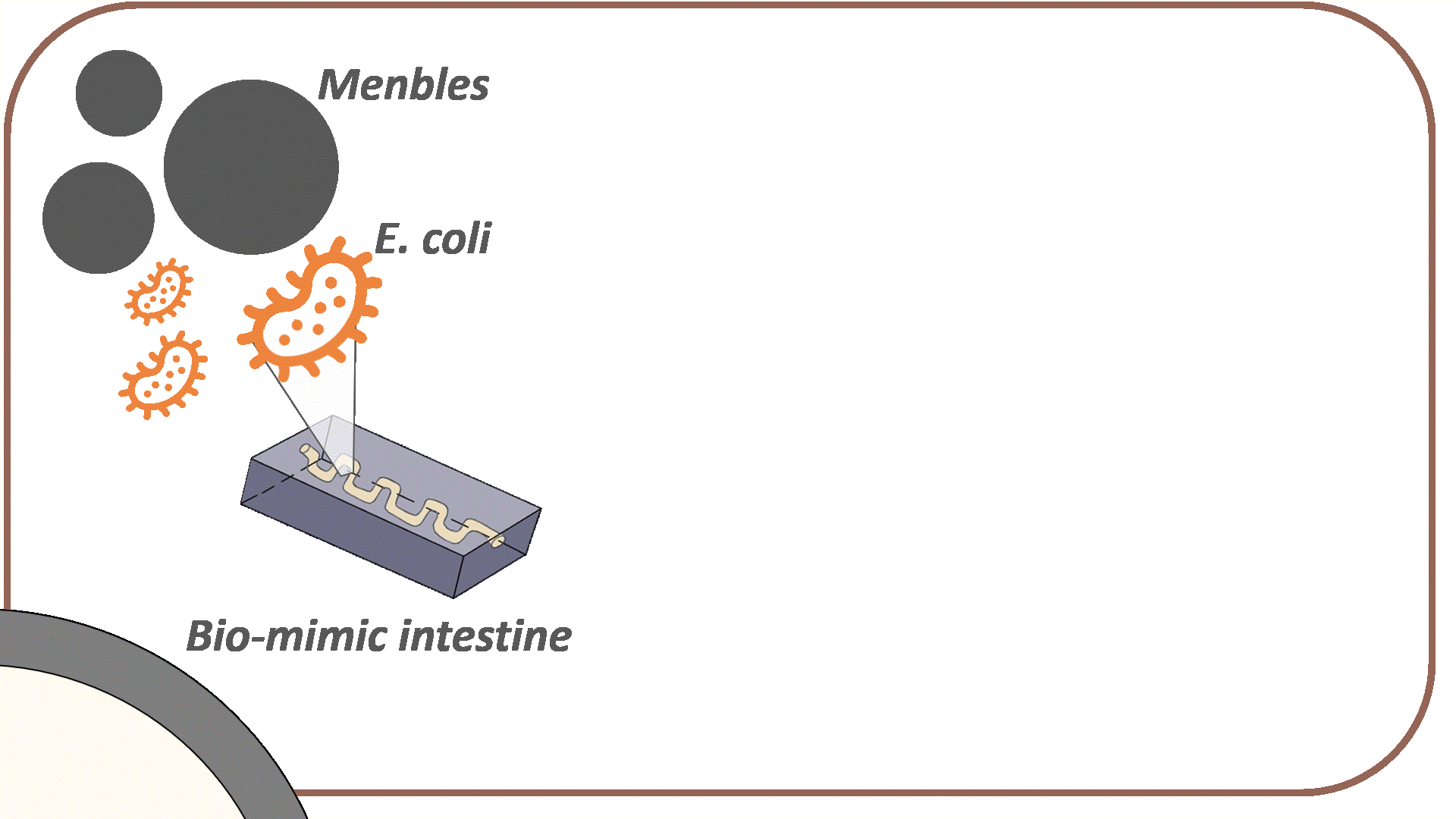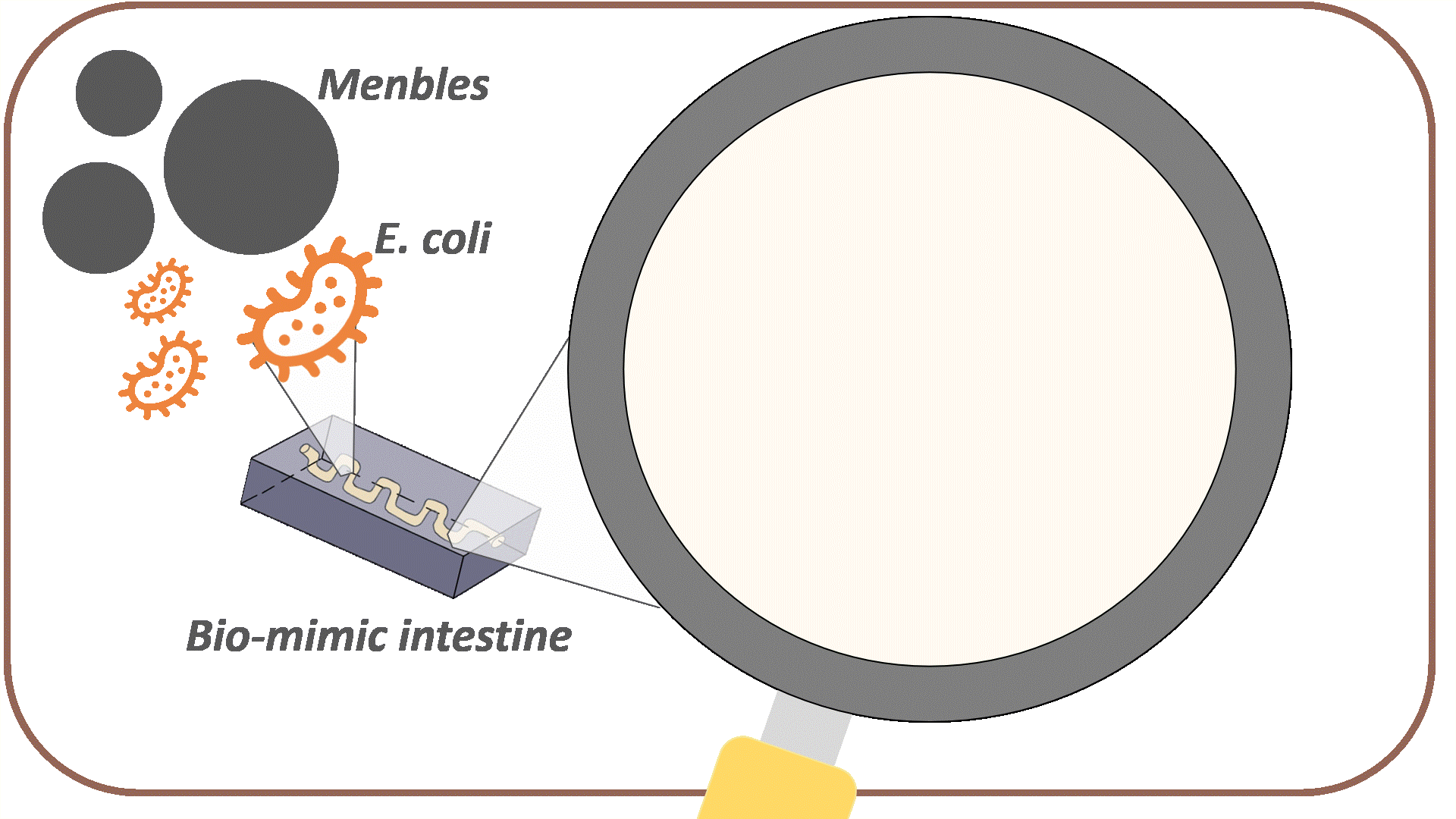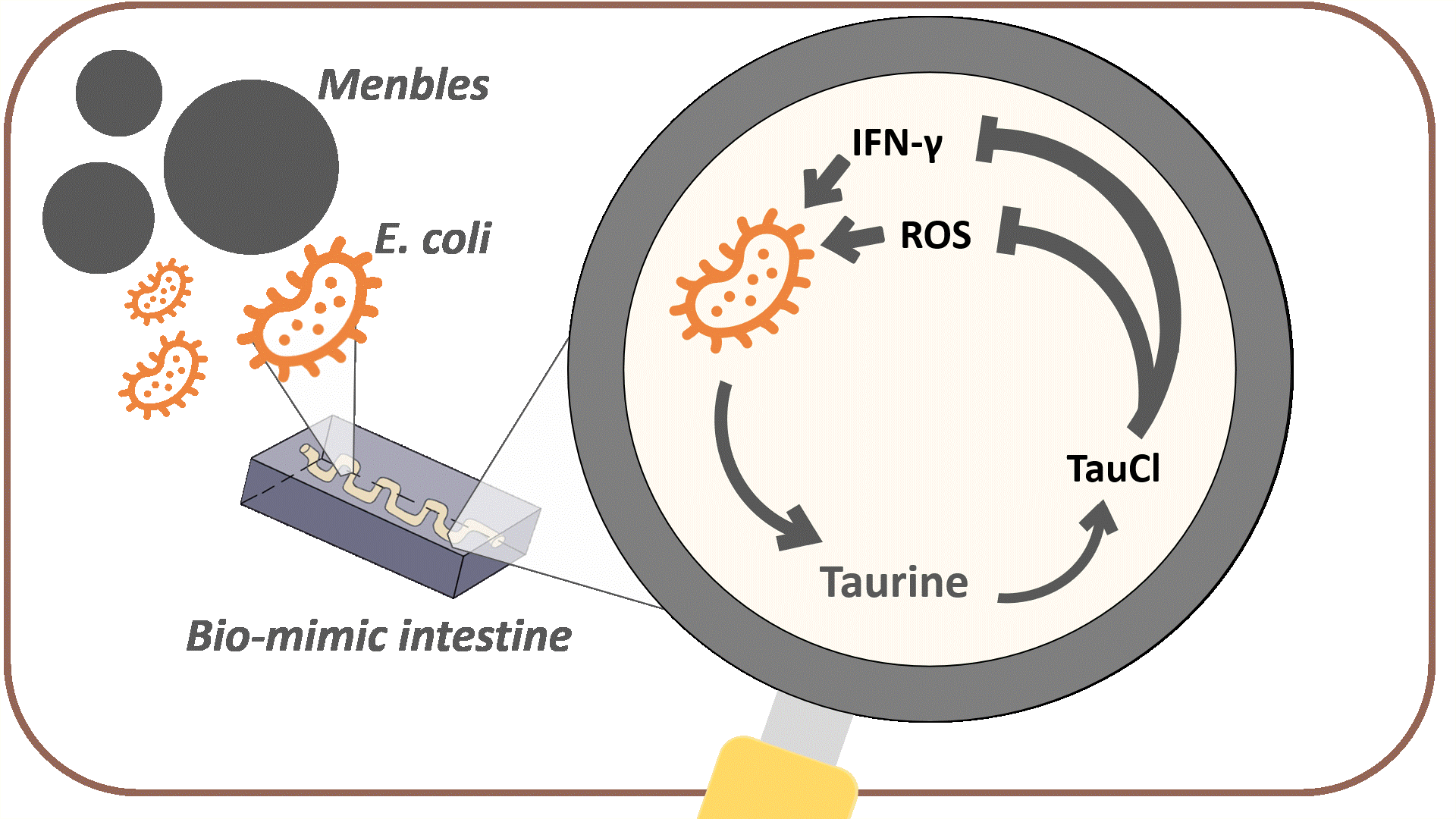
The overall goal of MenTAUR is to relieve depressive symptoms by increasing taurine levels in the intestine. To achieve this, we developed multiple models and conducted wet-lab experiments to prove that our bubble, Menbles, can effectively function and release taurine in the human intestine. Menbles are designed to withstand the acidic environment of the stomach, protecting our encapsulated bioengineered E. coli. We developed a microfluidic chip, f(int), that simulated the intestinal environment, allowing us to determine the retention of E. coli in the jejunum. After confirming that most of our E. coli can adhere to the intestinal walls, we proved that the E. coli could detect stress biomarkers reactive oxygen species (ROS) and IFN-γ through the oxidative stress and IFN-γ sensing systems and produce the taurine production enzymes. Finally, with math-based calculations, we can model the mechanism through which taurine converts into TauCl and reduces ROS and IFN-γ levels in the body, relieving depressive symptoms through the gut-brain axis.
Bubble Production
Menbles were special edible bubbles that serve as a unique depression medication, coming with a trendy drink worldwide, bubble milk tea. Menbles were designed to be incorporated into everyone’s daily lives, including people who cannot seek help due to stigma. They could quickly get their hands on this drink with functional bubbles.
The biggest challenge to Menbles is the digestion process in the stomach. Menbles must be ensured to withstand the presence of gastric acid in the stomach, as they were designed to release the bacteria in the intestine. Luckily, sodium alginate can withstand these extreme conditions. Sodium alginate has been proven to be resilient even under extreme stomach conditions[1]. Therefore, we believe it is ideal for our project to encapsulate our bacteria and not release it until it reaches the intestine, where combined action of acid and trypsin can break the alginate structure down[1], releasing the bacteria.
However, another challenge we encountered is that although alginate performs very well under acidic conditions, it does not perform too well under basic conditions[2]. Therefore, we tested our bubbles containing E. coli Nissle 1917 to be submerged into phosphate buffers of four different pHs, reflecting the range of pHs of milk tea. Furthermore, we tested different bubble sizes because some might experience difficulty swallowing the bubbles without chewing[3]. We were then inspired to design the bubbles as tiny as we could. Hence, we conducted experiments using different sizes of bubbles to identify which size has the best recovery rate for our Menbles. Below are the results of the rate of recovery of our bubble for the 14.14 mm3 (Large) bubble, 4.19 mm3 (Medium) bubble, and 1.77 mm3 (Small) bubble (Figure 2). For more detailed information, please refer to the Results page.
We hypothesized that as the pH goes higher, the bacteria content in the supernatant is higher due to the breaking down of alginate (in higher pH)[2]. Based on our results in Fig. 2, we concluded that the ideal size we would use for Menbles is the 4.19 mm3 (Medium) bubble because its results best fit our hypothesis. Besides, its bubble recovery is the most stable among other sizes.
Microfluidic Device
After we consume the engineered E. coli Nissle bubbles, there are almost no function or model that able to predict the number of probiotics remain in the human body accurately. As a result, we intended to take the retention ratio of E. coli Nissle in the jejunum into consideration. Nevertheless, due to the restriction of human trials and ethical issues, we came up with an idea to invent f(int) - a microfluidic channel to simulate the jejunum environment.

In our Generation 1 experimental results , as shown in Fig. 4, we found that the retention ratio of microfluidic chip w/ CF+2villi is higher than any other channel. In addition, when we compare the result of the same channel with additional 5% HA (Hyaluronic acid) that will be the substitution of mucus, the retention ratio increases by 5.45% (From 80.69% to 86.14%).

The approach that we took to use 5% HA is because jejunum intestinal juice has 0.2 to 5% mucin(MUC2)[4], and HA was used because of its same sticky characteristic with mucin and due to its low availability. Our experiment result shows that the microfluidic channel with a complex structure such as channel w/ CF+2villi that includes additional HA is the most suitable for simulating the human intestine.
Sensing for Stress Biomarkers
Oxidative Stress Sensing System Experiments
After confirming that most of the engineered bacteria can adhere to the walls of the intestine, we must prove that the bacteria can produce taurine. We designed our engineered bacteria to produce these enzymes only when the body is under high-stress levels, which are signaled by high levels of reactive oxygen species (ROS) and IFN-γ. The following experiments were performed to confirm that both oxidative stress and IFN-γ sensing systems can effectively detect their respective stress biomarkers and initiate taurine production.
The oxidative stress sensing system regulates the production of L-cysteine sulfinic acid decarboxylase (CSAD). It consists of the soxR gene (BBa_K3771100), promoter PsoxS (BBa_K3771048), and the csad gene (BBa_K3771008). When SoxR protein is fully oxidized, it becomes a powerful transcription activator of PsoxS, leading to the expression of the downstream gene. To test the intensity and specificity of the oxidative stress sensing system, the oxidative stress assay was performed.
Oxidative Stress Assay
Achievements
1. Compared the intensity and sensitivity of different promoters under the stimulation by oxidative stress.
2. Selected a suitable promoter for regulating the expression of L-cysteine sulfinic acid decarboxylase (CSAD).
Process
We compared the relative fluorescence intensity of sfGFP expressed by different promoters when induced by different concentrations of paraquat (PQ). Paraquat can react with oxygen to produce various types of reactive oxygen species (ROS), such as superoxide radical (O2−•), hydrogen peroxide (H2O2), and hydroxyl radical (OH−•)[5], which is used to simulate abnormal oxidative stress in the intestines of stressed individuals.
Over a period of 4.5 hours after paraquat was added, the relative fluorescence intensity was measured. J23100, a constitutive promoter, being the positive control, shows no difference in sfGFP expression level between the samples with and without paraquat. The activity of PsoxS (BBa_K3771048) is higher than that of PkatG (BBa_K3771047), but the activity is greatest when the SoxR transcription factor (BBa_K3771100) is added in the construction together with PsoxS (Fig. 6).
Another oxidative stress assay was performed only with SoxR and PsoxS promoter to determine the efficiency of this detection system. The sfGFP level was measured hourly during the 4.5 hours after paraquat induction, and results showed that even low concentrations of paraquat were able to activate the detection system (Fig. 8 and 9).
Interferon-gamma Sensing System Experiments
The second detection system is the IFN-γ sensing system, which regulates the production of CS.
The pathway for the IFN-γ sensing system requires two steps. First, ompA promoter constitutively expresses the OmpA/OprF chimeric protein, which is located in the plasma membrane of E. coli. Then, IFN-γ binds to the chimeric protein, activating a series of signal transduction reactions and finally inducing the pspA promoter and producing the CS enzyme. An IFN-γ induction assay was performed to confirm the function of the IFN-γ sensing system.
IFN-γ Induction Assay
Achievements
1. Selected a suitable promoter for regulating the expression of chimeric OmpA/OprF protein
2. Determined the minimum IFN-γ concentration required for activation of the sensing system
3. Determined the time required for the system to produce a significant difference in protein expression
Process
The two plasmids PpspA-ofp-PlacI-ompA/oprF (BBa_K3771017) and PpspA-ofp-PompA-ompA/oprF (BBa_K3771018) were transformed into ompA gene knockout strain, JW0940, to prevent OmpA from interfering with IFN-γ binding to chimeric OmpA/OprF. Human IFN-γ of concentrations 300 pM and 600 pM were added to bacteria cultures to induce the system. OFP expression (RFU) was recorded using a microplate reader and normalized by OD600. As shown in Fig. 11 and 12, starting at around 12 hours after induction, OFP expression of both samples increased dramatically.
As presented in Fig. 13, at 12 hours after induction, sample 2 (PpspA-ofp-PompA-ompA/oprF) (BBa_K3771018) with the addition of 600 pM IFN-γ had higher OFP expression than that of sample 1 (PpspA-ofp-PlacI-ompA/oprF) (BBa_K3771017). Since ompA promoter stimulates higher protein expression than lacI promoter, we decided to incorporate the ompA promoter in our final biobrick to regulate the expression of cs.
Model
Since reducing the amount of ROS was the main goal of Menbles, the mechanism we proposed was the anti-oxidative characteristic of the taurine derivative, TauCl. TauCl could induce lots of antioxidants to be produced within the intestinal epithelial cell. We modeled how GSH, one of the main antioxidants, could relieve ROS (see Fig. 14) in a faster and stronger way. Followed by the physiological mechanism proposed in Description a healthy gut could bi-directionally influence the brain, ultimately reducing various depressive symptoms.
References
- Guo L, Goff HD, Xu F, et al. The effect of sodium alginate on nutrient digestion and metabolic responses during both in vitro and in vivo digestion process. Food Hydrocolloids. 2020;107:105304. doi:10.1016/j.foodhyd.2019.105304
- Chuang J-J, Huang Y-Y, Lo S-H, et al. Effects of pH on the Shape of Alginate Particles and Its Release Behavior. International Journal of Polymer Science. 2017;2017:1-9. doi:10.1155/2017/3902704
- Aldridge KJ, Taylor NF. Dysphagia is a Common and Serious Problem for Adults with Mental Illness: A Systematic Review. Dysphagia. 2011;27(1):124-137. doi:10.1007/s00455-011-9378-5
- Leal J, Smyth HDC, Ghosh D. Physicochemical properties of mucus and their impact on transmucsal drug delivery. International Journal of Pharmaceutics. 2017;532(1):555-572. doi:10.1016/j.ijpharm.2017.09.018
- Lascano R, Muñoz N, Robert G, et al. Paraquat: an oxidative stress inducer. Herbicides-properties, synthesis and control of weeds. 2012:135-148.