Overview
In the wet lab, we took into account the fermentation characteristics of chassis cells——the “Crabtree effect” and “diauxic shift”, which have important impacts for the global regulation of gene expression and prevent cell factory from converting carbon sources into target products at maximum efficiency. Therefore, SCUT-China want to construct hybrid-promoters for different carbon sources respond via regulatory elements of promoters related to the diauxic shift. Then it is expected that we will obtain a series of hybrid-promoters with different intensity. More importantly, we will apply the best one to the Nootkatone pathway finally, so as to enhance our yeast cell factory.
“Crabtree effect” and “Diauxic shift”
After constructing the Nootkatone synthesis pathway in Saccharomyces cerevisiae, we learned that competition for common pool of ATP, cofactors, coenzymes and precursors between native and heterologous pathways can adversely affect growth, lengthen fermentation, and decrease product yields. One way to avoid this problem is to activate the production pathway after the major growth phase[1]. But how we find the trigger that can activate the shift?
Through research, we found two interesting phenomena of S. cerevisiae[2, 3]:
“Crabtree effect” : When fermentable hexoses (e.g. glucose, fructose) are used as a carbon source, even under aerobic conditions, S. cerevisiae firstly ferments sugars rapidly into ethanol.
“Diauxic shift” : When the preferred sugar depletes, growth slows while appropriate metabolic networks are switched on for using an alternative carbon source (such as the ethanol previously produced). The cells then continue growth on the alternative substrate(s).

That is, a good regulatory trigger would be the diauxic shift following fermentation in typical Saccharomyces cerevisiae fermentation.
More excitingly, promoters are regulatory elements that combine with transcription factors that are activated under specific conditions to regulate gene expression in different time and space[4].
However, several commonly used promoters in yeast cell factories belong to glycolytic genes,highly expressed in actively growing yeast when glucose is used as a carbon source. But their expression can be suboptimal when alternate carbon sources are used, owing to lacking the desired induction properties[5, 6].
Thus, it would be beneficial for our cell factory efficiency to using synthetic promoters with optimal regulatory output at the diauxic shift, which would be helpful to our Nootka-boom!

Carbon source response mechanism of promoter

S. cerevisiae promoters have multiple essential elements for the accurate transcriptional regulation of genes, including a core promoter region, an upstream activator sequence (UAS), an upstream repressor sequence (URS) and nucleosome-disfavoring sequences. The upstream activating sequence (UAS) is located upstream of the core promoter and serves as a binding site for specific transcription activators[7]. Different types of carbon sources have different activation effects on cells, making them express different transcription activators. Certain transcription activators bind to UASs of promoters, recruit RNA polymerase, and regulate genes globally.

To achieve gene expression differences under different carbon sources. Different UASs on each promoter give it the ability to respond to different carbon sources[8]. Therefore, we can recombine the UASs from different promoters to change promoters, making them adapt for different carbon sources and activate production in the diauxic shift automatically, namely our magical hybrid-promoters!
Promoter engineering
(1)Firstly, we need to know which of the saccharomyces cerevisiae endogenous promoters involved in glucose metabolism is most effective for expressing Valencene synthase and has the greatest potential for modification. 14 yeast promoters with potential in different metabolic pathways (glucose degradation, gluconeogenesis and pentose phosphate pathways) were found through literature review[9]. We chose TPI1p(Triose phosphate isomerase), TDH1p ( Glyceraldehyde-3-phosphate dehydrogenase), ENO1p(Enolase I), CDC19p(Cell Division Cycle/ Pyruvate kinase)、 PDC1p(Pyruvate Decarboxylase 1), PDC6p(Pyruvate Decarboxylase 6)in the Glycolysis pathway. Promoters of the Ethanol metabolic pathway include: ALD4p(Aldehyde Dehydrogenase), ALD3p(Aldehyde Dehydrogenase);The promoters of Pentose Phosphoate pathway include SOL4p (Suppressor of Los1-1/ 6-phosphogluconolactonase), ZWF1p(Glucose-6-phosphate dehydrogenase), TKL2p(Transketolase).
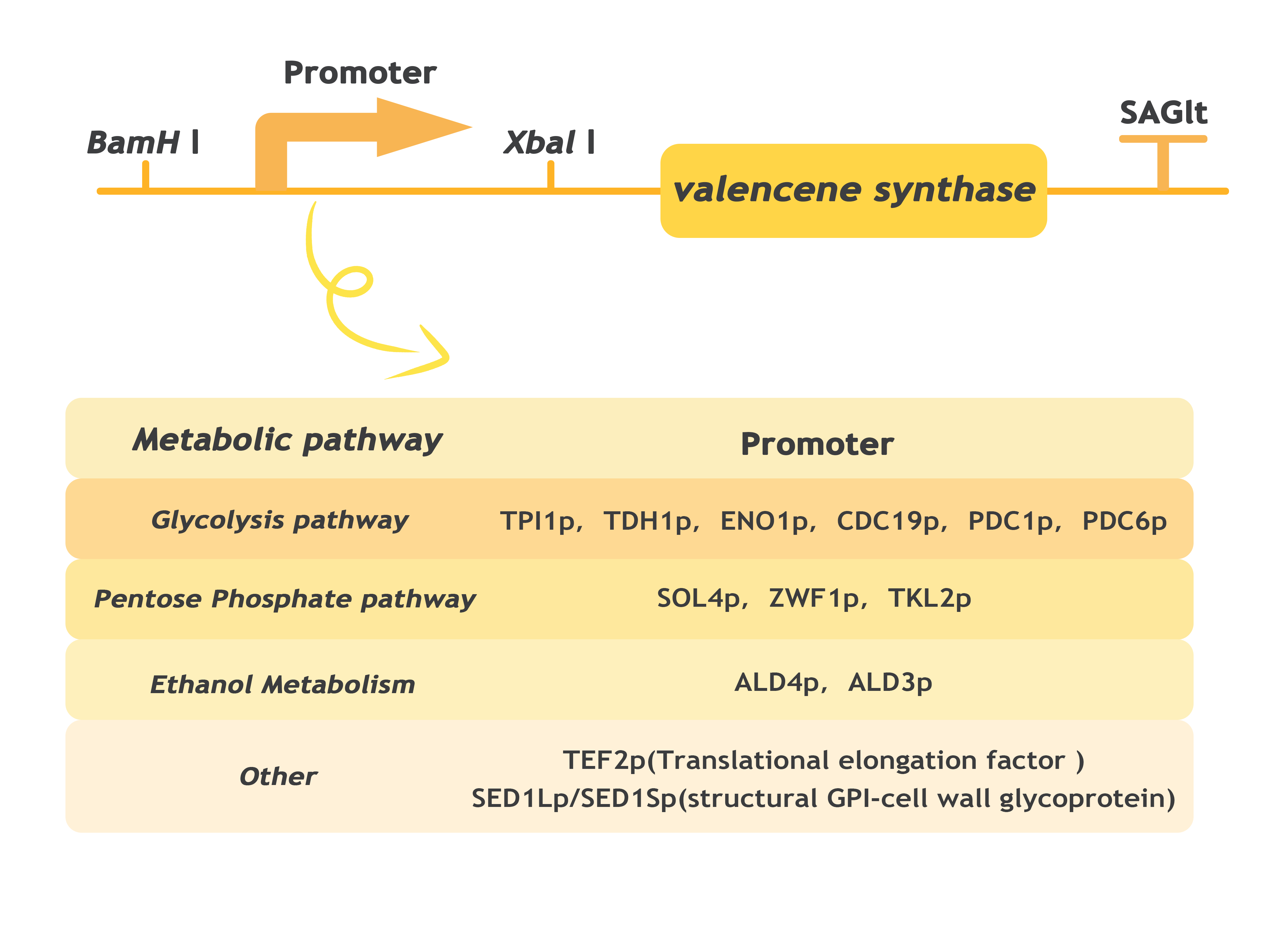
Next, we constructed Valencene expression cassettes containing different promoters. With CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats , we knocked the expression cassettes into the genome of S. Cerevisiae BJ5464. and high intensity promoters were screened by shaker fermentation and gas chromatography.

The results showed that PPDC1 was the promoter with the best expression effect of Valencene Synthase, which would be used as the original promoter for our subsequent modification.

(2)Secondly, we selected high-strength promoters from different metabolic pathways, namely, PDC1p of glycolysis pathway, ALD4p of ethanol metabolism pathway, SED1Lp of cell wall structure gene, and ZWF1p of pentose phosphate pathway. By comparing the time curve and specific rate curve of Valencene production, the characteristics of the promoter were investigated.

From the figures below (figure 1&2), we have learned key information and further understood the reasons behind: PPDC1 is mainly expressed in the glucose consumption stage (0-16h), and valencene production reaches a stable level at 24h, indicating that PPDC1 is a promoter that regulates the synthesis of valencene in the early stage. PALD4 is strictly limited in controlling valencene synthesis and is not expressed during glucose utilization by cells and is only effective when ethanol is consumed. ALD4 encodes mitochondrial aldehyde dehydrogenase, which catalyzes acetaldehyde to acetate, and is an important part of ethanol utilization pathway in S. cerevisiae[10]. When S. cerevisiae uses ethanol, the enzyme will be activated. The strength of SED1Lp is weak relatively during glucose consumption and increases significantly at the late fermentation stage (32-56 h). SED1 encodes structural proteins in the cell wall that are highly expressed during quiescence and protect cells from environmental stresses such as heat, ethanol, and lyase[11]. When cells are at rest, SED1 will activate transcription. The overall expression level of PZWF1 is low, so it is not considered to be used for our subsequent promoter engineering.


The production rate of valencene can more directly reflect the expression characteristics of pPDC1, pSED1L, pALD4, pZWF1.

Based on the promoter characteristics obtained from the above experimental results in the design stage, we will subsequently modify the transcription factor binding site of promoter candidate PPDC1 in the engineering part to enhance the strength of the promoter at the later stage of fermentation.
References
1.Rajkumar, A.S., et al., Engineered Reversal of Function in Glycolytic Yeast Promoters. Acs Synthetic Biology, 2019. 8(6): p. 1462-1468.
2.Peng, B., et al., Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: a comparison of yeast promoter activities. Microbial Cell Factories, 2015. 14.
3.Lee, K.M. and N.A. DaSilva, Evaluation of the Saccharomyces cerevisiae ADH2 promoter for protein synthesis. Yeast, 2005. 22(6): p. 431-440
4.Hahn, S. and E.T. Young, Transcriptional Regulation in Saccharomyces cerevisiae: Transcription Factor Regulation and Function, Mechanisms of Initiation, and Roles of Activators and Coactivators. Genetics, 2011. 189(3): p. 705-736.
5.Partow, S., et al., Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast, 2010. 27(11): p. 955-964.
6.Weinhandl, K., et al., Carbon source dependent promoters in yeasts. Microbial Cell Factories, 2014. 13.
7.Tang, H., et al., Promoter Architecture and Promoter Engineering in Saccharomyces cerevisiae. Metabolites, 2020. 10(8)
8.Cao, L., et al., Two-stage transcriptional reprogramming in Saccharomyces cerevisiae for optimizing ethanol production from xylose. Metabolic Engineering, 2014. 24: p. 150-159.
9.Nambu-Nishida, Y., et al., Selection of yeast Saccharomyces cerevisiae promoters available for xylose cultivation and fermentation. Journal of Bioscience and Bioengineering, 2018.125(1): p. 76-86.
10.Navarro-Avino, J.P., et al., A proposal for nomenclature of aldehyde dehydrogenases in Saccharomyces cerevisiae and characterization of the stress-inducible ALD2 and ALD3 genes. Yeast, 1999. 15(10A): p. 829-842.
11.Shimoi, H., et al., Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. Journal of Bacteriology, 1998. 180(13): p. 3381-3387.


