
Overview
We verified in engineering success that CARγ macrophages(Mγ) and CARMERTK(Ms) have the phagocytic capacity specific for SARS-COV-2 and showed pro-inflammatory and anti-inflammatory properties respectively. And we designed our target patients in proposed implementation. This is the silver medal work that we have done. Based on the above, we did the following work to verify that Toggle Macrophage can eliminate virus and regulate immune level in human body according to our expected work.
1. Designed and tested IL-6 Sensor and gene circuit.
2. Fully considered the safety problems that may be encountered in clinical practice.
3. Establish a model to prove that the number of Toggle Macrophage, N, has a certain range [a,b] to make IL-6 oscillate around a certain stable value M when the initial value of IL-6 concentration is i0, and the range of oscillation is [l,u].
A concern of phenotypic regulatory role of Macrophages
After coming up with our Using envision, we communicated closely with doctors and immunologists who had worked on the front lines of the fight against the pandemic. They asked the following three questions (more details in Integrated HP )that prompted us to design and test Toggle Macrophage to demonstrate that our project is likely to work in a relevant context.
Question1: In the early stages of infection, the body's natural immune response helps to clear the virus due to a low viral load. Imagine if clinicians misjudge the use of CAR-M therapy, it will suppress the immune response of the body, resulting in the worsening of the disease, and the body can not properly perform an effective immune response.
Improvement: We constructed the proinflammatory CAR-γmacrophages (Mγ), to address the problem of low immunity in the early stage of COVID-19 infection. (See the CAR γsection in Design for details ) And in vitro experiments have verified that M γ promotes the secretion of cytokines and strengthens the body's elimination of viruses. (Click Engineering Success: Testing the CAR receptors for details)
Question2: After talking with Professor Dong Yuchao, who went to the front line of the fight against COVID-19, we learned that the symptoms and cytokine levels of COVID-19 patients did not exactly match during the clinical diagnosis process, and they could not accurately judge the timing of the application of the therapy. Even when serum cytokine concentrations are measured at any time, transient changes in inflammation levels cannot be obtained.
Improvement: We decided to integrate CARMERTK and CARγ, and design a Smart macrophage, which regulates the phenotypic transformation between Mγ and Ms through a cytokine sensor, so as to realize virus clearance and bidirectional regulation of immunity. (See Circuit Design for details )
Question3: There are a variety of cytokines in COVID-19 patients. Which cytokines should be selected as indicators of CRS?
Improvement: We reviewed the literature on IL-6 signaling pathway and designed the IL-6 cytokine sensor after several rounds of brainstorming. We hope that by designing IL-6 receptor, the influence of IL-6 on macrophages will be expanded far beyond that of other cytokines.
Through gene circuit design, we successfully verified in vitro experiments that IL-6 Sensor activated GFP (representing CARγ expression) and mCherry (representing CARMERTK expression) respectively according to the concentration of IL-6 in the environment.
Design of IL-6 sensor and linked gene Circuits
Cytokines and chemokines overproduced in a cytokine storm include IL-1β, IL-2, IL-6, IL-10, TNF-α, IFN-γ, IP-10, MIP-1, and MIP-1α (Cao X et al.2020). IL-6 is thought to be a key factor, because serum IL-6 concentration was associated with disease severity (Velavan TP et al.2020) and mortality (Ruan Q et al.2020).

IL-6 is synthesized by macrophages and dendritic cells upon recognition of pathogens through toll-like receptors at the site of infection. Circulating IL-6 binds to the soluble form of IL-6 receptor, forming a complex with a gp130 dimer on potentially all cell surfaces. The complex activates the JAK-STAT3 signaling pathway in various cell types, resulting in CRS and finally leading to ARDS (He X et al.2020). Blocking this pathway means performing a therapeutic effect against cytokine release syndrome (CRS) in patients with a severe form of COVID-19.
Drugs that block IL-6 activity has been proved effective. For example, treatment with tocilizumab, a monoclonal antibody that binds the soluble form of IL-6 receptor, markedly improved the respiratory condition and resolved computed tomography findings of the lungs in 80% of severely ill COVID-19 patients (Xu X et al.2020).
We therefore constructed IL-6 sensor on the CAR-expressing macrophages, serving as an essential button to determine whether to initiate the transformation.

According to the in vitro experiments, CARγ macrophage (Mγ) can promote inflammation and CARMERTK macrophage (Ms) can inhibit inflammation. We determined to integrate CARγ and CARMERTK into a single macrophage by our circuit design in order to construct the smart Toggle Macrophage which can respond to the changing microenvironment by switching its phenotype. The circuit we designed determines the function of Toggle Macrophage by regulating the CAR expression based on the IL-6 concentration in the microenvironment. The circuit is as followed:
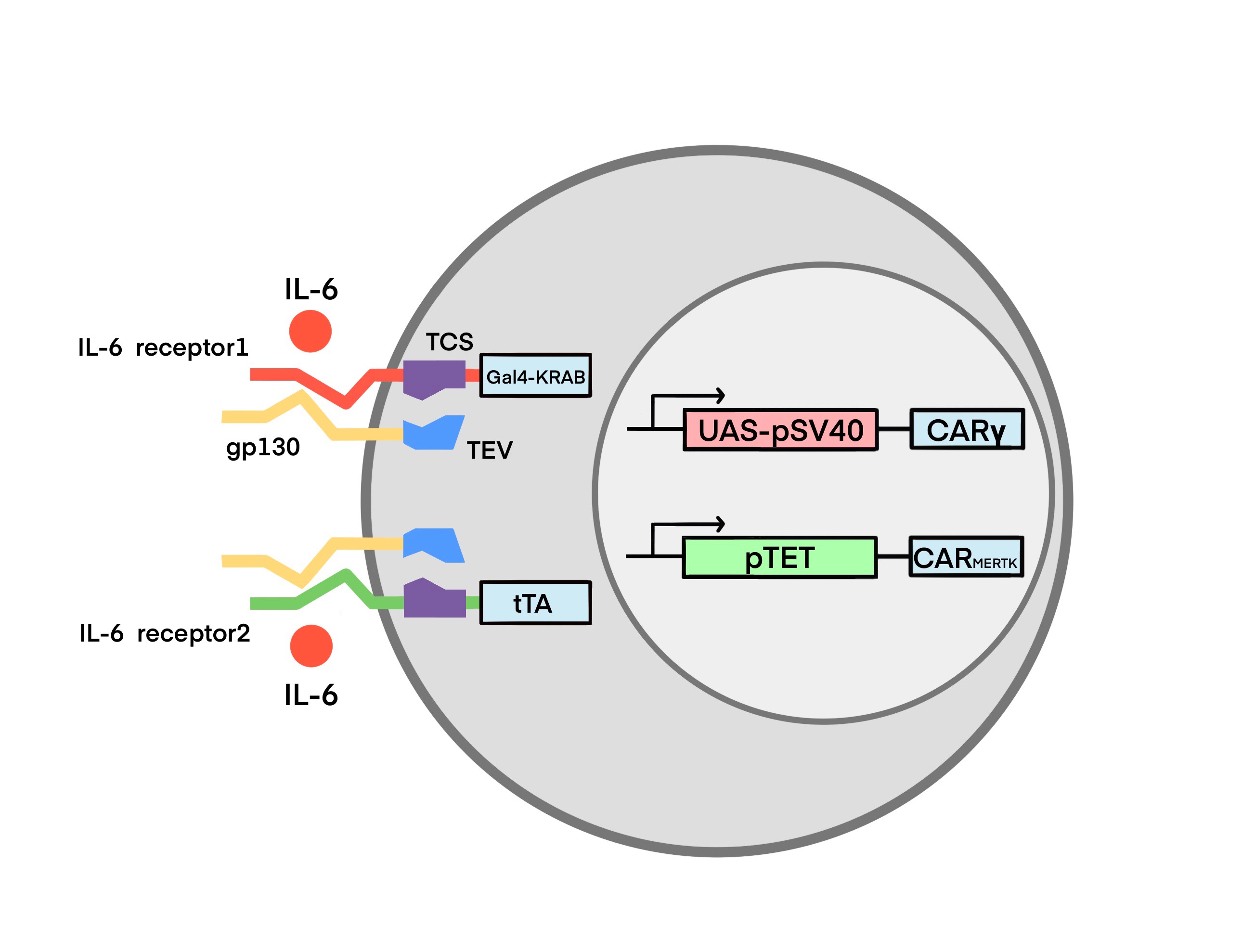
The Structure of IL-6 Sensor
Based on the high affinity binding of IL-6 to IL-6R and gp130 expressed by monocytes(Hunter and Jones 2015). We designed the IL-6 receptor with two parts. The first part consists of a membrane-type gp130 with tobacco etch virus protease (TEVp). TEVp is a highly specific cysteine protease in TEV. A number of TEVp mutants with different rate of cleavage, stability and specificity have been reported. Similarly, a panel of different TEV cleavage sites(TCS), derived from the canonical ENLYFQ-G/S site, has been established. TEVp is highly valued for its good transduction efficiency and specificity. To our knowledge, it was never reported that TEVp would cleave fusion proteins out of target. Moreover, TEVp harbors several advantages in biotechnological applications, such as simple production process, low cost, and utility of open source vectors and mutants, making it suitable for different in vitro and in vivo applications(Cesaratto, Burrone, and Petris 2016).

The second part consists of the extracellular domain of membrane IL-6R, TEV cleavage site (TCS) and transcription factors. The transcription factors are the transcriptional repressor GAL4-KRAB (IL-6 receptor 1) and the transcriptional activator tTA (IL-6 receptor 2), respectively. Specific information about GAL4-KRAB and tTA will be described in the Module 2.


When the IL-6 concentration in the environment reaches a certain threshold, IL-6 receptors are activated as follows: (Ps: For both receptors, the triggering process is the same)
1.IL-6 binds to membrane IL-6R (mIL-6R) with low affinity.
2. IL-6-IL-6R complex binds to gp130 extracellularly.
3.TEV protease downstream of gp130 is activated and hydrolyzes the TCS.
4. The transcription factor Gal4-KRAB (IL-6 receptor 1)/tTA (IL-6 receptor 2) is released.
Linked Gene Circuits of IL-6 Receptor 1: (GAL4-KRAB-UAS-pSV40- CARγ)

GAL4-KRAB:
This part is a mammalian synthetic transcription factors based on Gal4 DNA binding domain (DBD)(Pengue et al. 1994) and KRAB transcription repression domain. Gal4-KRAB containing three core domains from N-terminal to C-terminal: GAL4 DNA binding domain, nuclear location sequence (NLS) and KRAB transcription repression domain(Morsut et al. 2016). And a (G4S) linker was added between DBD and NLS for providing region flexibility(Witzgall et al. 1994; Chen, Zaro, and Shen 2013). GAL4 DBD is capable of binding to specific DNA sequences and KRAB repressing the expression of downstream gene, so that we used Gal4-KRAB as a transcription repression factor to inhibit the activation of downstream synthetic promoter UAS-pSV40.
1.KRAB(Kruppel-associated box): KRAB is the N-terminal reserve area of Kruppel-associated protein, Kid-1, which can reversibly inhibit the gene expression by forming heterochromatin via recruiting various histone mediation factors(Wang et al. 1997). The KRAB protein has been demonstrated to be capable of inhibiting all promoters within at least 3 kB(Deuschle, Meyer, and Thiesen 1995). Strong transcriptional repression was observed when the KRAB domain was bound both at near or kilobase distances from the start site of transcription.
2.Gal4: Gal4 is a yeast transcriptional activator consisting of 881 amino acid. The DNA binding activity of Gal4 is located in the 74 amino acids in the N terminus (Keegan et al. 1986). The transcriptional activation function of Gal4 is mapped in two regions (residues 148–196 and 768–881) (Ma & Ptashne 1987). Gal4 binds to its specific recognition sequence UAS (upstream activating sequence) and activates transcription of target genes. It has been demonstrated that the Gal4-UAS system can operate not only in yeast but also in various animal cells(Fischer et al. 1988).
UAS-pSV40-CARγ:
1.GAL4 upstream activation sequence (UAS): In cells where the Gal4 gene is expressed, the Gal4 protein targets the UAS sequence(Guarente, Yocum, and Gifford 1982), thus driving expression of any open reading frame immediately downstream of the UAS. In principle, this allows for any gene (linked to UAS) to be expressed in any pattern, so long as there is a known enhancer driving Gal4(Scott 2009; Wang et al. 1997).
2.SV40 promoter (pSV40): the pSV40 is expressed well in the fission yeast S. pombe, and it initiates transcription at the same site as in mammalian cells. In mammalian cells, SV40 early gene transcription is predominantly initiated from a set of closely adjacent sites 24-27 bp downstream of the TATA sequence(Benoist and Chambon 1981)
3.aSscFV-Fcγ(CAR γ)

tTA (tetR-VP64):
tTA contains three core domains from N-terminal to C-terminal: DNA-binding transcriptional repressor (tetR), NLS and VP64 transcription activation domain. And a linker was added between DBD and NLS for providing region flexibility. tetR DBD is capable of binding to specific DNA sequences and VP64 activating the expression of downstream gene, so that we used tetR-VP64 as transcription activation factor to activate our downstream synthetic cis-acting elements and promotor PminCMV.
1.Tetracycline repressor (tetR): tetR can negatively regulate the transcription of resistance-mediating genes in E.coli. In the presence of the antibiotics tetracycline or doxycycline (Dox), tetR does not bind to its operators (tetO) located within the promoter region of the operon and allows transcription(Gossen and Bujard 1992). In the absence of the antibiotics, tetR binds to tetO and thus represses transcription. Herein, tetR acts as the DBD of tTA, which can specifically bind to TRE (fused with seven tetO)(Gossen and Bujard 1992).
2.VP64: VP64 is a fusion of four VP16, which is more potent than VP16. VP16 is an activating factor that primes transcription from the five virally encoded immediate early (IE) genes in HSV-1(Triezenberg, Kingsbury, and McKnight 1988). The natural tetR is a transcriptional repressor protein in E. coli. Fusing VP64 to tetR can transform tetR can be modified into a transcriptional activator protein available in eukaryotes(Gossen and Bujard 1992). Herein, VP64 acts as the transcription activation domain (AD) of tTA.
pTET (TRE-PminCMV)-CARMERTK:
1.TRE: TRE is the fusion of seven tetracycline operators (tetOs). tetO can be bound with tetR. When bound to tetOs placed upstream of minimal promoters, tTA efficiently activates transcription from such promoters(Gossen and Bujard 1992).
2.PminCMV: PminCMV downstream of TRE is a CMV promoter lacking an enhancer termed minimal CMV promoter, which is inefficient in initiating transcription and can be approximated as being unable to initiate transcription without the combination of tetR and tetO(Gossen and Bujard 1992).
3.aSscFV-MERTK (CARMERTK):

Illustration of Tet-off system. pTET (BBa_K1061013) consists of TRE and PminCMV. Tandem TetO sequences are positioned upstream of the PminCMV followed by cDNA of gene of interest. Here, a chimeric protein consisting of TetR and VP64 (tTA) is converted into a transcriptional activator, and the expression plasmid is transfected together with the operator plasmid. Thus, culturing cells with Dox switches off the exogenous gene expression, while removing Dox switches it on(Kallunki et al. 2019).
Through the circuit design, we aimed to achieve the following design:

As described above, neither IL-6 receptor is triggered when the IL-6 concentration in the body does not reach a threshold, and the transcription factors Gal4-KRAB (IL-6 receptor 1) and tTA (IL-6 receptor 2) are not released. The following effects occur.
a. IL-6 receptor 1 does not trigger: initiating CARγ transcription
1. pSV40 promoter activates transcription of the gene aSscFV-Fcγ.
2. CARγ is abundantly expressed on the cell membrane.
b. IL-6 receptor 2 does not trigger: repressing CARMERTK transcription
1. PminCMV is highly inefficient and fails to initiate transcription.
2. CARMERTK is barely expressed on the cell membrane
As can be seen, the inactivation of IL-6 receptor 1 and 2 results in enhancing CARγ expression and suppressing CARMERTK expression, respectively. In general, these effects promote the conversion of toggle macrophages into Mγ. The proportion of Mγ in the macrophage cluster gradually rises and results in phagocytosing the viruses and enhancing the immune function of the patient.

As described above, when the IL-6 concentration within the environment reaches a threshold, both IL-6 receptors are triggered and the transcription factors Gal4-KRAB (IL-6 receptor 1) and tTA (IL-6 receptor 2) are released. The following effects are produced.
c. IL-6 receptor 1 triggers: suppressing CARγ transcription
1. Gal4 binds to UAS.
2. KRAB represses the expression of the downstream gene aSscFV-Fcγ.
d. IL-6 receptor 2 triggers: promoting CARMERTK transcription
1.tetR binds to tetO.
2.VP64 binds to the corresponding cis-acting elements.
3.transcription of the gene aSscFV-MERTK is then initiated by PminCMV downstream of TRE.
The activation of IL-6 receptor 1 and 2 results in repressing CARγ expression and promoting CARMERTK expression, respectively. In general, these effects promote the conversion of toggle macrophages into Ms. The proportion of Ms in the macrophage cluster gradually rises and results in phagocytosing the viruses and suppressing the cytokine storm.

Ms then causes a gradual decrease in IL-6 concentration, the transcription of CARγ gene restarts, while the transcription of CARMERTK gene gradually stops. Correspondingly, the Ms gradually apoptosis, while the Mγ gradually proliferate, and finally the macrophage cluster reassumes the function of Mγ again, forming an effective negative feedback loop regulatory mechanism. Ultimately, we hope to reach a ‘cytokinostasis’, keeping the IL-6 concentration within a safe range through the intelligent conversion of macrophages.

Obtaining Parts
For IL-6 Sensor and linked circuit, we have designed 10 basic parts synthesized by GENEWIZ and then we used these basic parts to synthesize 6 composite parts. We also gained 7 basic parts and 3 composite parts from:
| Name | Type | Description |
|---|---|---|
| BBa_K4040006 | Protein_Domain | TEV protease |
| BBa_K4040011 | Protein_Domain | tTA |
| BBa_K4040013 | Protein_Domain | mIL-6R |
| BBa_K4040014 | Protein_Domain | CD8a signal peptide |
| BBa_K4040019 | Protein_Domain | CR3022 scFv |
| BBa_K4040032 | Protein_Domain | IL-6 signal transducer, gp130 |
| BBa_K4040034 | Protein_Domain | Truncated HER2 |
| BBa_K4040035 | Protein_Domain | Truncated EGFR |
| BBa_K4040036 | Protein_Domain | CD20 |
| BBa_K4040037 | Protein_Domain | T2A peptide |
| Name | Type | Description |
|---|---|---|
| BBa_K4040020 | Composite | GP130-TEV |
| BBa_K4040021 | Composite | IL6R-TCS-Gal4-KRAB |
| BBa_K4040022 | Composite | IL6R-TCS-tTA |
| BBa_K4040023 | Composite | pSV40-UAS-GFP Expression Cassette |
| BBa_K4040024 | Composite | pTET-mCherry-PGK-CD20 Expression Cassette |
| BBa_K4040033 | Composite | UAS-pSV40 |
| Name | Type | Description |
|---|---|---|
| BBa_J18918 | Protein_Domain | TEV cleavage site |
| BBa_I712004 | Regulatory | CMV promoter |
| BBa_E0840 | Reporter | GFP |
| BBa_J04450 | Reporter | MCherry |
| BBa_K3064008 | Regulatory | PGK |
| BBa_K1061011 | Regulatory | EF-1alpha promoter |
| BBa_K1061013 | Regulatory | P tight promoter |
| BBa_K2446037 | Composite | ZF_GAl4_KRAB |
| BBa_K511003 | Composite | pSV40_UAS |
| BBa_K1061013 | Composite | TRE- pmincmv |
Testing Overall circuit
1. Transduction screening In order to select the macrophages in which the genetic circuits have successfully expressed, we linked several parts which could be detected in each circuit. As shown in Figure 1,2,3 synthetic receptors contained the gp130-TEV, IL-6R- TCS -Gal4-KRAB and IL-6R- TCS- tTA. The cDNA sequences containing the various fusion constructs were cloned into the pELNS vector with the promoter of CMV.

Figure1

Figure2

Figure3
The reporter GFP ( BBa_E0840) was fused to the promoter UAS-pSV40 ( BBa_K511003). (figure 4) We used mCherry ( as the reporter and connected it to the pTET ( BBa_K1061013), followed by PGK promoter and CD20. (figure 5)

Figure4

Figure5
1e7 cells were co-transfected with the plasmid. As shown in Figure 6, we screened 1e7 cells after transduction to obtain cells successfully expressing five cassettes for subsequent experiments. EGFR and HER2 positive cells were screened first (in the box), and GFP and Myc positive cells were screened later (in the box). CD20 positive cells were then screened (in the box). The cells obtained were used in subsequent experiments to detect the effects of IL-6.

Figure6
2. Detection of IL-6 receptor effect
We added different concentrations of IL-6 to the above screened cells to detect mCherry and GFP fluorescence intensity, and the results were shown in Figure 7. MCherry fluorescence intensity represents pTET pathway expression, which is used to reflect CAR-MERTK expression in vivo. GFP fluorescence intensity represents the expression of UAS-PSV40 pathway, corresponding to the expression of CAR-γ in vivo. As can be seen from the figure7, CAR-Ms change from M1 to M2 when IL-6 concentration is 2.5-12.5ng/mL.

Figure7
Concern of Safety
In order to guarantee the safety of Toggle Macrophage in vivo, we have made some safety considerations.
We promise that we will not take any biological samples out of our BSL-2 laboratory,and our operators have accepted safety training. (See safety page for more information.)
Reliable source and expansion are the necessary conditions for the clinical application of CAR-M, which can be prepared by PBMC or by iPSCs. In addition, unlike T cells, macrophage has a low risk of GVHD, which means that products can be made in advance for patients to use on demand. (Chen et al., 2021)
CRS represents the major problem of CAR T therapies. Giulia’s analysis has not revealed new adverse drug reactions. (Giulia et al., 2021) Due to the Immunoregulation capability of Toggle macrophage, the possibility of cytokine storm is small.
As for Toggle Macrophage, we have demonstrated some safety aspects in vitro experiment.
1. Do not harm normal cells(Ms)
The phagocytic potential of human macrophage THP-1 cell lines expressing different CAR receptors or a truncated CAR receptor (CAR△) lacking the intracellular domain was measured with a cell-based assay. Consistent with previous reports (Morrissey et al., 2018), CAR macrophages and control untransduced (UTD) macrophages did not show notable phagocytosis of 293 cells; however, CARMEGF10, CARγ and CARz cells but not CARMERTK, CARD, or UTD macrophages phagocytosed Spike-bearing 293 cells in an S-specific manner (Figure 1).

Figure 8
CAR-mediated macrophage phagocytosis was further confirmed by a luciferase-based killing assay, and our data showed that CARMEGF10, CARg and CARz cells eradicated S protein expressing 293T cells in an antigen-specific manner (Figure 2). Interestingly, CARMERTK and UTD macrophages showed-no difference in killing effect.

Figure 9
The above two experiments suggest that CARMERTK macrophages (Ms) do not phagocytose or lyse 293T cells expressing S protein (Mimicking infected normal cells) under high inflammatory conditions. This feature is beneficial for COVID-19 patients suffering from cytokine storms, considering that the release of lysosomal inflammatory substances after cell membrane destruction leads to a strong inflammatory response. Of course, this property brings with it the problem of not being able to eliminate the infected normal cells, which is also a safety threat to patients. Our interpretation is that suppression of the inflammatory response is more urgent than elimination of the virus when cytokine storms occur. After the cytokine storm subsides, Ms switches to Mγ to quickly eliminate the virus.
2. Would not be infected
Although there is currently no evidence that SARS-CoV-2 can infect THP-1 cells with or without IgGs (Banerjee et al., 2020), THP-1 cells have been shown to support antibody-mediated enhancement of SARS-CoV infection in previous studies (Jaume et al., 2021). We therefore sought to determine whether synthetic receptors facilitate the entry of SARS-COV-2 into macrophages as host cells, as the extracellular domain of the CAR constructs has the capacity to directly bind to the S protein. We employed VSV pseudovirus bearing SARS-2-S (Kleine-Weber et al., 2019) to study the cell entry of SARS-CoV-2. Our data showed that Vero E6 cells were susceptible to entry driven by SARS-S (Figure 3); however, no evidence of infection was detected in THP-1 cells with or without synthetic receptors. Synthetic receptors did not facilitate the entry of SARS-COV-2 into macrophages as host cells.

Figure 10
Although Toggle Macrophage is considered safe in vivo, a final safety catch is still needed. This way, the Toggle Macrophage can be rapidly eliminated in vivo. An excellent candidate for this inducible kill switch is the AP1903 described by Clackson, et al., 1998, as AP1903 has no other biologic effects in vivo. (Straathof, K. C., et al.,2016) ICasp9 can be activated by AP1903 that has proven safe at the required dose for optimal eradicating effect. (Iuliucci, J. D., et al.,2001) (More details in Safety )

We have verified that Toggle Macrophage can regulate immune response in a bidirectional way by in vitro experiments. However, the environment in vivo is often complex. So we established a model to quantitatively analyze the cytokine levels that Toggle Macrophage can control. The model can tell us that the concentration range of IL-6 in patients can be greatly improved by importing a certain amount of macrophages, which greatly improves the safety of Toggle Macrophage.

Model for clinicians
As mentioned above, we have qualitatively verified this by experiment. The designed synthetic antiviral macrophages, termed "Toggle Macrophages", have the phagocytic capacity specific for SARS-CoV-2 and showed pro-inflammatory/anti-inflammatory phenotypic transformation according to the level of local IL-6 concentration. But as a novel Genetically Engineered Cellular Platform that may be useful in future therapeutic approaches against severe COVID-19, we need to carry out quantitative analysis by establishing models. (more details in Model)
Our model found the following patterns:
1. For a given initial value of IL-6 concentration i0, n has a certain range [a,b] in which IL-6 oscillates around a certain stable value M, and the range of oscillation is [L, U].
2. Within the range of [A, B], the larger the n value is, the larger the il-6 stable value m is, and the larger the oscillation range is, but the time required to reach the oscillation is shorter.
3. Within the range of [A, B], the smaller the n value is, the smaller the il-6 stable value m is, and the smaller the oscillation range is, but the time required to reach the oscillation is longer.
Therefore, we assume that clinicians measure the concentration of IL-6 in the serum of patients, and input the concentration of IL-6 in tissues calculated by our Formula (3) to obtain the range [A, B] of the number of macrophages n that can make IL-6 oscillate at a certain stable value M. In order to figure out the optimal number of macrophages n, we need to further limit the range of oscillation according to clinical requirements. Here, we propose the following constraints:
1. The high value U of oscillation is not higher than the IL-6 concentration of severe patients.
2. ......
There are many clinical limitations on cytokine levels, and we just gave some examples. In conclusion, a feasible number of macrophages can be determined by further limiting the oscillation range [A,B].
Reference
Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol 2020; 20: 269-270 [PMID: 32273594 DOI: 10.1038/s41577-020-0308-3]
Velavan TP, Meyer CG. Mild versus severe COVID-19: Laboratory markers. Int J Infect Dis 2020; 95: 304-307 [PMID: 32344011 DOI: 10.1016/j.ijid.2020.04.061]
Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med 2020; 46: 846-848 [PMID: 32125452 DOI: 10.1007/s00134-020-05991-x]
He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, Mo X, Chen Y, Liao B, Chen W, Hu F, Zhang Q, Zhong M, Wu Y, Zhao L, Zhang F, Cowling BJ, Li F, Leung GM. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 2020; 26: 672-675 [PMID: 32296168 DOI: 10.1038/s41591-020-0869-5]
Benoist, C., and P. Chambon. 1981. 'INVIVO SEQUENCE REQUIREMENTS OF THE SV40 EARLY PROMOTER REGION', Nature, 290: 304-10.
Cao, Xuetao. 2020. 'COVID-19: immunopathology and its implications for therapy', Nature Reviews Immunology, 20: 269-70.
Cesaratto, Francesca, Oscar R. Burrone, and Gianluca Petris. 2016. 'Tobacco Etch Virus protease: A shortcut across biotechnologies', Journal of Biotechnology, 231: 239-49.
Chau, A. S., A. G. Weber, N. I. Maria, S. Narain, A. Liu, N. Hajizadeh, P. Malhotra, O. Bloom, G. Marder, and B. Kaplan. 2021. 'The Longitudinal Immune Response to Coronavirus Disease 2019: Chasing the Cytokine Storm', Arthritis Rheumatol, 73: 23-35.
Chen, Xiaoying, Jennica L. Zaro, and Wei-Chiang Shen. 2013. 'Fusion protein linkers: Property, design and functionality', Advanced Drug Delivery Reviews, 65: 1357-69.
Choi, B. D., X. L. Yu, A. P. Castano, A. A. Bouffard, A. Schmidts, R. C. Larson, S. R. Bailey, A. C. Boroughs, M. J. Frigault, M. B. Leick, I. Scarfo, C. L. Cetrulo, S. Demehri, B. V. Nahed, D. P. Cahill, H. Wakimoto, W. T. Curry, B. S. Carter, and M. V. Maus. 2019. 'CAR-T cells secreting BiTEs circumvent antigen escape without detectable toxicity', Nature Biotechnology, 37: 1049-+.
Deuschle, U, W K Meyer, and H J Thiesen. 1995. 'Tetracycline-reversible silencing of eukaryotic promoters', Molecular and Cellular Biology, 15: 1907-14.
Edelstein, H. I., P. S. Donahue, J. J. Muldoon, A. K. Kang, T. B. Dolberg, L. M. Battaglia, E. R. Allchin, M. Hong, and J. N. Leonard. 2020. 'Elucidation and refinement of synthetic receptor mechanisms', Synth Biol (Oxf), 5: ysaa017.
Fischer, Janice A., Edward Giniger, Tom Maniatis, and Mark Ptashne. 1988. 'GAL4 activates transcription in Drosophila', Nature, 332: 853-56.
Giavridis, Theodoros, Sjoukje J. C. van der Stegen, Justin Eyquem, Mohamad Hamieh, Alessandra Piersigilli, and Michel Sadelain. 2018. 'CAR T cell-induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade', Nature Medicine, 24: 731-+.
Gossen, M., and H. Bujard. 1992. 'TIGHT CONTROL OF GENE-EXPRESSION IN MAMMALIAN-CELLS BY TETRACYCLINE-RESPONSIVE PROMOTERS', Proceedings of the National Academy of Sciences of the United States of America, 89: 5547-51.
Guarente, L., R. R. Yocum, and P. Gifford. 1982. 'A GAL10-CYC1 HYBRID YEAST PROMOTER IDENTIFIES THE GAL4 REGULATORY REGION AS AN UPSTREAM SITE', Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences, 79: 7410-14.
Hao, Zhaonian, Ruyuan Li, Li Meng, Zhiqiang Han, and Zhenya Hong. 2020. 'Macrophage, the potential key mediator in CAR-T related CRS', Experimental Hematology & Oncology, 9.
Hunter, Christopher A., and Simon A. Jones. 2015. 'IL-6 as a keystone cytokine in health and disease', Nature Immunology, 16: 448-57.
Kallunki, Tuula, Marin Barisic, Marja Jäättelä, and Bin Liu. 2019. 'How to Choose the Right Inducible Gene Expression System for Mammalian Studies?', Cells, 8: 796.
Liu, Yili, Daihong Chen, Junjie Hou, Haicong Li, Dan Cao, Mingquan Guo, Yun Ling, Menglu Gao, Yi Zhou, Yanmin Wan, and Zhaoqin Zhu. 2021. 'An inter-correlated cytokine network identified at the center of cytokine storm predicted COVID-19 prognosis', Cytokine, 138.
Morrissey, M. A., A. P. Williamson, A. M. Steinbach, E. W. Roberts, N. Kern, M. B. Headley, and R. D. Vale. 2018. 'Chimeric antigen receptors that trigger phagocytosis', Elife, 7.
Morsut, Leonardo, Kole T. Roybal, Xin Xiong, Russell M. Gordley, Scott M. Coyle, Matthew Thomson, and Wendell A. Lim. 2016. 'Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors', Cell, 164: 780-91.
Pengue, Gina, Viola Calabró, Paola Cannada Bartoli, Alfredo Pagliuca, and Luigi Lania. 1994. 'Repression of transcriptional activity at a distance by the evolutionary conserved KRAB domain present in a subfamily of zinc finger proteins', Nucleic Acids Research, 22: 2908-14.
Rafiq, S., C. S. Hackett, and R. J. Brentjens. 2020. 'Engineering strategies to overcome the current roadblocks in CAR T cell therapy', Nature Reviews Clinical Oncology, 17: 147-67.
Reeh, H., N. Rudolph, U. Billing, H. Christen, S. Streif, E. Bullinger, M. Schliemann-Bullinger, R. Findeisen, F. Schaper, H. J. Huber, and A. Dittrich. 2019. 'Response to IL-6 trans- and IL-6 classic signalling is determined by the ratio of the IL-6 receptor alpha to gp130 expression: fusing experimental insights and dynamic modelling', Cell Commun Signal, 17: 46.
Roybal, K. T., J. Z. Williams, L. Morsut, L. J. Rupp, I. Kolinko, J. H. Choe, W. J. Walker, K. A. McNally, and W. A. Lim. 2016. 'Engineering T Cells with Customized Therapeutic Response Programs Using Synthetic Notch Receptors', Cell, 167: 419-32 e16.
Schwarz, K. A., N. M. Daringer, T. B. Dolberg, and J. N. Leonard. 2017. 'Rewiring human cellular input-output using modular extracellular sensors', Nature Chemical Biology, 13: 202-09.
Scott, Ethan K. 2009. 'The Gal4/UAS toolbox in zebrafish: new approaches for defining behavioral circuits', Journal of Neurochemistry, 110: 441-56.
Tan, P., L. He, G. Han, and Y. B. Zhou. 2017. 'Optogenetic Immunomodulation: Shedding Light on Antitumor Immunity', Trends in Biotechnology, 35: 215-26.
Tang, Yueting, Jiayu Sun, Huaqin Pan, Fen Yao, Yumeng Yuan, Mi Zeng, Guangming Ye, Gui Yang, Bokun Zheng, Junli Fan, Yunbao Pan, Ziwu Zhao, Shuang Guo, Yinjuan Liu, Fanlu Liao, Yongwei Duan, Xiaoyang Jiao, and Yirong Li. 2021. 'Aberrant cytokine expression in COVID-19 patients: Associations between cytokines and disease severity', Cytokine, 143.
Todt, J. C., B. Hu, and J. L. Curtis. 2004. 'The receptor tyrosine kinase MerTK activates phospholipase C gamma 2 during recognition of apoptotic thymocytes by murine macrophages', Journal of Leukocyte Biology, 75: 705-13.
Triezenberg, S. J., R. C. Kingsbury, and S. L. McKnight. 1988. 'Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression', Genes Dev, 2: 718-29.
Wang, Y., J. Xu, T. Pierson, B. W. Omalley, and S. Y. Tsai. 1997. 'Positive and negative regulation of gene expression in eukaryotic cells with an inducible transcriptional regulator', Gene Therapy, 4: 432-41.
Witzgall, R., E. O'Leary, A. Leaf, D. Onaldi, and J. V. Bonventre. 1994. 'The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression', Proceedings of the National Academy of Sciences of the United States of America, 91: 4514-8.
Wu, C. Y., K. T. Roybal, E. M. Puchner, J. Onuffer, and W. A. Lim. 2015. 'Remote control of therapeutic T cells through a small molecule-gated chimeric receptor', Science, 350: aab4077.
