
Description
Background
At present, food corruption has become a major issue in food safety. Among them, the problems caused by fish spoilage are particularly obvious. According to incomplete statistics, there are more than 200 species of diseases in aquaculture nationwide, among which dozens are common diseases. The pathogens include viruses, bacteria, fungi, and parasites, bacterial diseases accounted for 57.63%.In 2018, the economic loss of China's aquaculture caused by diseases was about 45 billion RMB (The data is from the internet).
The spoilage rate of fish is relatively fast, and it will spoil soon after being caught (within about 12 hours). Fish are more susceptible to spoilage mainly due to the following reasons: 1. The method of obtaining aquatic products: Most fish die when they are caught. After the fish is caught and killed, they no longer have the ability to resist the invasion of microorganisms, and microorganisms can invade fish tissues in different ways. 2. The transportation and storage process of aquatic products: microorganisms often invade fish tissues from mechanical wounds caused by fishing, fish storage, and transportation. 3. The problem of the fish itself: the fish body has a high water content (about 70%-80%) and relatively rich protein content, fragile tissues, and fish scales are easy to fall off, causing bacteria to easily invade from the injured part, and the mucus on the surface of the fish body is a good culture medium for bacteria, so microorganisms can invade fish tissues from the mucus of the epidermis, and the gills that are most susceptible to bacterial invasion.

Fig.1 Fish storage
Theoretically, live fish tissue is sterile, however, the living environment is not. So there are a certain number of microorganisms on the surface, gills, and digestive tract of the fish. After transportation, storage, and processing, the types and number of microorganisms on the surface of the fish may increase. Generally speaking. The bacteria that marine fish carry and can cause fish spoilage are common Pseudomonas, Achromatic chrysanthemum, Flavobacterium, Morseella, etc. These microorganisms use the mucus on the surface of the fish as a medium to grow and multiply and secrete a large number of proteases to decompose the protein in the fish meat so that the fish meat epidermal tissue changes from hard to loose till entire fish muscle tissue is destroyed (Tang et al.,2000).


Fig.2. Spoilaged fish (left) and Pseudomonas Aeruginosa(right)
(Spoilage fish picture was from Souhu internet)
With the booming fishery industry, consumers are increasingly demanding fresh fish every year. Whether in the process of farming, transportation, or storage, spoilage caused by microbial invasion will affect the fish itself and the health of consumers. Pseudomonas Aeruginosa is an important pathogenic bacteria of fish, which can lead to high mortality of fish and corruption of fish and fish products (Abdullahi et al., 2013), which may lead to health problems of consumers. Therefore, real-time detection of P. aeruginosa can help aquaculture plants deal with corrupt fish in time. It can also help consumers confirm whether they are edible or not before eating. However, a timely and convenient detection method is still lacking at present.
Currently methods
In order to detect the bacteria, there are several ways to deal with it. First of all, sampling is directly used for culture detection of bacteria, which is time-consuming and can only be limited to the laboratory. Since the quorum-sensing signal system of P. aeruginosa has been well studied, the detection of its signal molecules can also be used to detect the bacterium. P. aeruginosa has four typical QS signal systems(figure 3), among which las and rhl systems have been studied most deeply and extensively. These two signaling systems are involved in the synthesis of N-(3-oxododecanoyl) homoserine lactone (3O-C12-HSL) and (C4-HSL) signaling molecules(figure 4), respectively. The existing detection methods of 3O-C12-HSL and C4-HSL mainly include LC-MS/MS, HPLC-MS/MS, Dual Signal Confirmation at Microelectrode Arrays, etc. These methods have some problems such as long measuring time, high cost, and strict requirements on experimental equipment.

Fig 3.Quorum sensing system of Pseudomonas Aeruginosa


Fig 4. Chemical Structure of C4-HSL(left) and 3O-C12-HSL(right)
So there is an increasing need for a simple, portable, and accurate product that could be used by anyone who needs to detect fish spoilage under any circumstances. According to the public survey conducted by us, 72.94% of people think that detecting fish spoilage in time is very important, thus, it inspires us that we can design such a product and promote it to the masses.
Our solution
Our project aims to utilize the aptamer-based lateral flow method to detect 3O-C12-HSL and C4-HSL. The method has a variety of advantages such as high efficiency, time, sensitivity, and simplicity. It is easy to use and can be applied by anyone in any scenario. More importantly, it has a relatively cheaper price, which makes it can be used in a wider range compared to current methods.
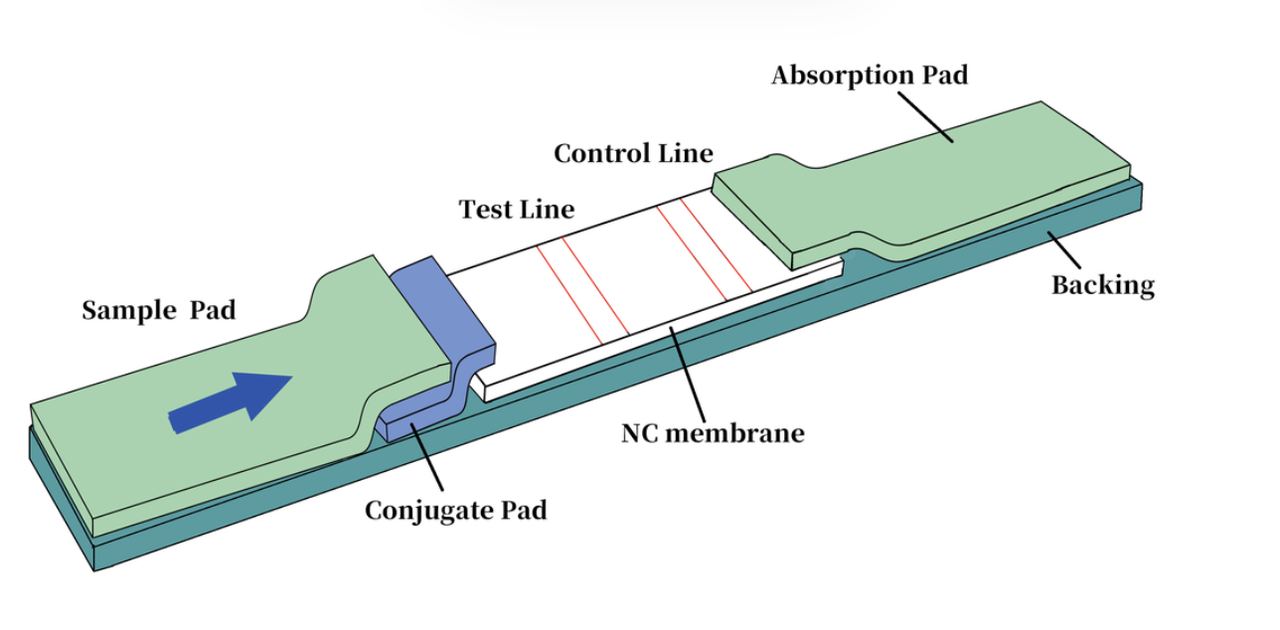
Fig 5. The design of aptamer-based lateral flow assay
What's more, we would like to construct a system that can detect 3O-C12-HSL and C4-HSL based on the transcription factor, namely the receiver cell that can sense the autoinducer signal and characterize it by a reporter gene. We believe the final product would be different from the aptasensor. The two kinds of biosensors combined can be applied to a wider range and more operating systems. Although at last due to the time and epidemic limit, we only constructed the aptasensor successfully, we found a receiver cell in a paper that can sense the C4-HSL and used GFP as the reporter gene, we acquired the E.coli from the author(Du et al., 2020)and test it, then combined it with hydro system design by NDNF_China, we accomplished another biosensor.
Our design
We would use the aptamers that can be specifically bound to 3O-C12-HSL and C4-HSL found in papers and verify the function by excitation and quenching of SG (SYBR GreenⅠ). We also used the FAM-labeled and BHQ-labeled anti-aptamer to test the aptamer, which was another fluorescence method. In the process of the experiment, we also rationally designed the aptamers in order to make them have a better binding ability to our target molecules. Finally, we gained our aptamers with a high affinity to targets that could be used in the product design (McKeague et al., 2013; Li et al., 2018).
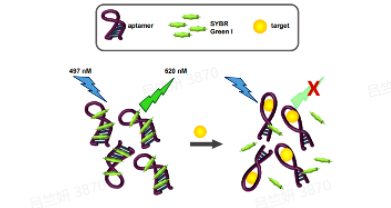

Fig 6.The fluorescence method of SG (left) and FAM (right)(McKeague et al., 2014)
See our Proof_Of_Concept and Engineering success
Then, the aptamers were used in the construction of aptamer-based lateral flow assay, and our final product was designed. Thus, we improved the detection 3O-C12-HSL and C4-HSL with less time consuming and price. A suitable method could be provided to the public(figure 7).

Fig 7. The 3D model of our product
See ourHardware and Implementation
At the same time, we conducted a series of human practices communications, providing suggestions and supports for every stage of our project. More importantly, we hold some educational activities, we hope that more people, especially middle school students of our age, will understand the origin, goals, development status, and prospects of this subject. At the same time, our team hopes to cultivate the scientific spirit of the new generation, establish a correct scientific view, and explain bioethics to them. This is also the purpose of our activities.
See our Communication and Human_Practices
We also did some modeling based on our experiment, in the SG fluorescence method, we firstly used the model to fit the combination mechanism of SG and aptamer, then we used the binding and dissociation model to and the experimental data of C4-HSL and aptamer, finally, we acquired the KD(equilibrium dissociation constant) value of the aptamer, proving the affinity of our aptamers to targets.
See our Model
References:
- Abdullahi, R. et al., Detection of oprL gene and antibiotic resistance of Pseudomonas aeruginosa from aquaculture environment. Euro. J. Exp. Bio,2013.3(6): p. 148-152.
- Du, P., et al., De novo design of an intercellular signaling toolbox for multi-channel cell-cell communication and biological computation. Nat Commun, 2020. 11(1): p. 4226.
- Li, Y., L. Sun, and Q. Zhao, Development of aptamer fluorescent switch assay for aflatoxin B1 by using fluorescein-labeled aptamer and black hole quencher 1-labeled complementary DNA. Anal Bioanal Chem, 2018. 410(24): p. 6269-6277.
- McKeague, M., et al., Selection and characterization of a novel DNA aptamer for label-free fluorescence biosensing of ochratoxin A. Toxins (Basel), 2014. 6(8): p. 2435-52.
- Wang, L., et al., An aptamer-based chromatographic strip assay for sensitive toxin semi-quantitative detection. Biosens Bioelectron, 2011. 26(6): p. 3059-62.
- Zhou, W., et al., An aptamer based lateral flow strip for on-site rapid detection of ochratoxin A in Astragalus membranaceus. J Chromatogr B Analyt Technol Biomed Life Sci, 2016. 1022: p. 102-108.
- 唐裕芳.鱼肉的腐败机理及其防腐措施[J].肉类工业,2000(02):30-32.

