
Our Project, Collagene, concerns the production of bacterial collagen in yeast, in replacement of current extraction methods of animal collagen. For more details on how this is done, please reference our Project Design page for a more specific overview of our lab work and details on our lab procedures/protocol.
Project modeling is integral to gaining a better understanding of Collagene. Our modeling included two major aspects, genetic and molecular modeling of bacterial collagen, as well as mathematical modeling of our bioreactor.
Protein Modeling
Producing stable and structurally sound bacterial collagen is integral to our project’s success. To ensure that the strengths and qualities of the animal collagen are not lost in our replacement, we began our search for past literature with protein sequences of bacterial collagen that had been already built and utilized them to model the quality and strength. We found a protein sequence of a bacterial collagen-like protein document in a previous paper (Xu et al 2002). Using the protein BLAST software, we chose 100 homologous sequences.
Using the MUSCLE (Multiple Sequence Comparison by Log-Expectation) multiple sequence alignment algorithm provided by the MEGAX software, the homologous sequences were narrowed down in preparation for ancestral reconstruction.

Ancestral reconstruction is a commonly used technique to find and apply important traits that may have been lost in certain genes throughout evolution. The ANCESCON software, developed by the Grishin Lab, is able to reconstruct ancestral protein sequences, with consideration for variation of evolutionary rates between different positions in the evolution of protein families (Cai et al 2004). The software produced a phylogenetic tree to represent the ancestral reconstruction (Figure 2) and we used the following node weighting method developed by University of Exeter’s 2019 team:


Using this node-weighting model, we chose one suitable ancestor to analyze for structural validity. Using the SWISS-MODEL, a fully automated protein homology-modelling server, as well as the PyMol molecular visualization tool, where we looked for a variety of structural aspects. Past literature elucidates two main structural elements that are important for stability in bacterial collagen, a globular N-terminal domain, as well as a triple-helix structure (Mohs et al 2007).



In Figure 3, the triple helical structure of the bacterial collagen-like ancestor protein is clearly highlighted in different colors. Bacterial collagen is lacking in the amino acid hydroxyproline, which is found in animal collagen and promotes thermal stability. However, previous literature has found that a triple helical structure in bacterial collagen similar to the one modeled exhibits high thermal stability of up to 39°C (Yu et al 2014), much similar to the structure found in our chosen protein. The globular N-terminal trimerization domains, shown in the ancestral sequence in Figures 4.1, 4.2, have been shown in previous literature to be integral to the folding initiation and nucleation of the triple-helix structure, and therefore also to the thermal stability of the overall protein (Cheng et al 2011). The stability of the protein structure is largely dependent on both the triple helical structure and the N-terminal trimerization domains, and the models in Figures 3, 4.1, and 4.2 show the structures as both fundamental to the protein. Having verified the validity and feasibility of a bacterial collagen protein sequence, we continued with our lab work.
Bioreactor Modeling

In Figure 5.3, a graph of the cell density in our single chamber bioreactor was measured at various levels of agitation, which helps understand how high agitation was more effective at increasing cell growth. Considering the data from figure 5.4 which helped us understand that high agitation was most effective in causing yeast growth, our bioreactor was designed with quad propellers that spin at 350rpm. The shape of the propellers allows for maximum movement of yeast media, correlating with better yeast growth.
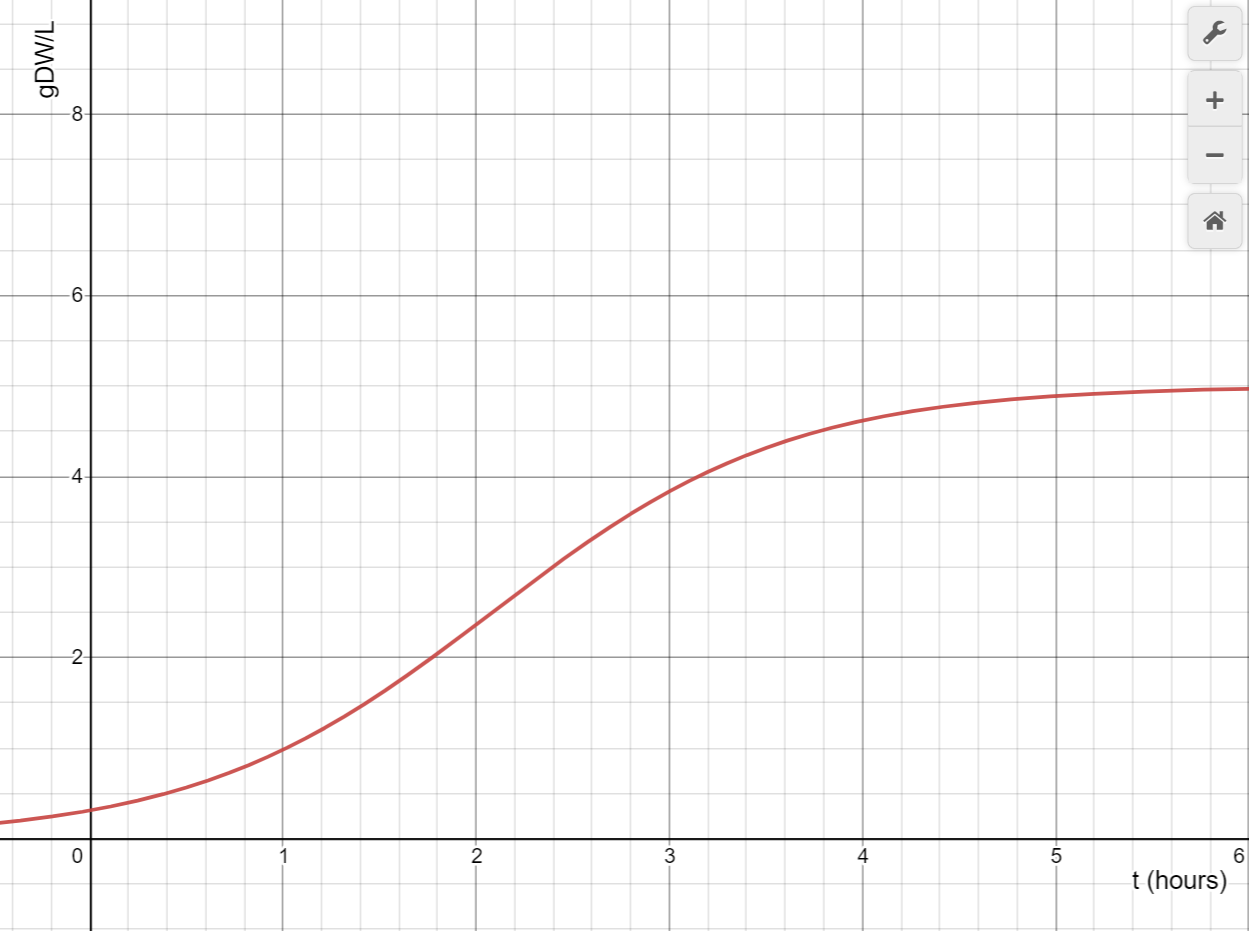

In Figure 5.4, a graph and equation were produced to estimate the change in yeast density for a 20 inch wide bioreactor. The single chamber design allows for a relatively small chamber to lead to the most effective production and cost efficiency. This allows the change in yeast density to be estimated, which can be helpful for determining the most efficient parameters for the bioreactor.
References
Cai, W., Pei, J. & Grishin, N.V. Reconstruction of ancestral protein sequences and its applications. BMC Evol Biol 4, 33 (2004). https://doi.org/10.1186/1471-2148-4-33
Lukomski, S., Bachert, B.A., Squeglia, F. and Berisio, R. (2017), Collagen-like proteins of pathogenic streptococci. Molecular Microbiology, 103: 919-930. https://doi.org/10.1111/mmi.13604
Mohs A, Silva T, Yoshida T, Amin R, Lukomski S, Inouye M, Brodsky B. Mechanism of stabilization of a bacterial collagen triple helix in the absence of hydroxyproline. J Biol Chem. 2007 Oct 12;282(41):29757-65. doi: 10.1074/jbc.M703991200. Epub 2007 Aug 10. PMID: 17693404.
Xu Y, Keene DR, Bujnicki JM, Höök M, Lukomski S. Streptococcal Scl1 and Scl2 proteins form collagen-like triple helices. J Biol Chem. 2002 Jul 26;277(30):27312-8. doi: 10.1074/jbc.M201163200. Epub 2002 Apr 25. PMID: 11976327.
Yu, Z., An, B., Ramshaw, J. A. M., & Brodsky, B. (2014). Bacterial collagen-like proteins that form triple-helical structures. Journal of Structural Biology, 186(3), 451–461. doi:10.1016/j.jsb.2014.01.003









