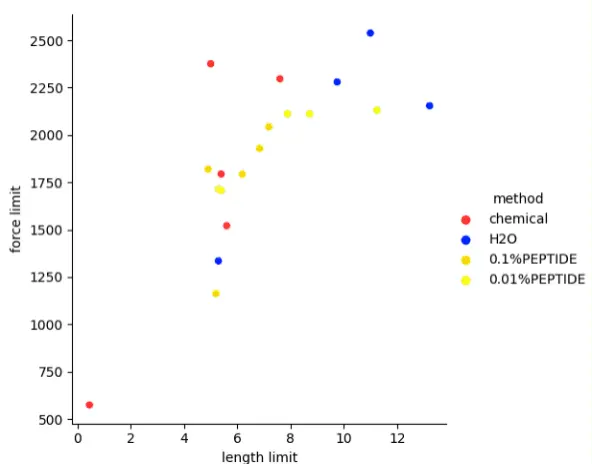
| − | <!-- # TODO: #6 Fix table caption font--><!-- # TODO: #7 Fix citations links font size--><html lang="en"><head><meta charset="utf-8"/><meta content="width=device-width,initial-scale=1" name="viewport"/><link href="https://2021.igem.org/Template:HUST-China/css/contentCSS?action=raw&ctype=text/css" rel="stylesheet"/><title>Results | iGEM HUST-China</title><script>MathJax={tex:{inlineMath: [['$', '$'], ['\(', '\)']],packages: {'[+]': ['mhchem']},loader: {load: ['[tex]/mhchem']},};</script><link href="https://2021.igem.org/Template:HUST-China/css/contentCSS?action=raw&ctype=text/css" rel="stylesheet"/></head><body><!-- # TODO: #6 Fix table caption font--><!-- # TODO: #7 Fix citations links font size--><nav class="navbar navbar-expand-xl fixed-top"><div class="container d-flex justify-content-between"><a class="navbar-brand" href="https://2021.igem.org/Team:HUST-China"><i class="navbar-logo-left"></i><span>HUST-China</span></a><button aria-controls="navbarNav" aria-expanded="false" aria-label="Toggle navigation" class="navbar-toggler" data-target="#navbarNav" data-toggle="collapse" type="button"><span class="navbar-toggler-icon"></span></button><div class="collapse navbar-collapse" id="navbarNav"><ul class="navbar-nav ml-auto"><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarProjectDropdown" role="button">Project</a><div aria-labelledby="navbarProjectDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Description">Description</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Design">Design</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Proof_Of_Concept">Proof of Concept</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Safety">Safety</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Implementation">Proposed Implementation</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Contribution">Contribution</a></div></li><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarHuman PracticesDropdown" role="button">Human Practices</a><div aria-labelledby="navbarHuman PracticesDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Human_Practices">Overview</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Human_Practices_01">Part 1</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Human_Practices_02">Part 2</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Collaborations">Part 3</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Education">Part 4</a></div></li><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarModelingDropdown" role="button">Modeling</a><div aria-labelledby="navbarModelingDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Model">Overview</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Gene_Pathway">Gene Pathway</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Dye_color_forecast">Dye color forecast</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Micronucleus_Counter">Micronucleus Counter</a></div></li><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarProductsDropdown" role="button">Products</a><div aria-labelledby="navbarProductsDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Products">Overview</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Software">Software</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Hardware">Hardware</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Another_Area">Another Area</a></div></li><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarExperimentsDropdown" role="button">Experiments</a><div aria-labelledby="navbarExperimentsDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Experiments">Overview</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Parts">Parts</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Engineering">Engineering Success</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Results">Results</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Protocol">Protocol</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Notebook">Notebook</a></div></li><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarPeopleDropdown" role="button">People</a><div aria-labelledby="navbarPeopleDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Team">Team</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Partnership">Partnership</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Attributions">Attributions</a></div></li><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarAchievementsDropdown" role="button">Achievements</a><div aria-labelledby="navbarAchievementsDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Judging">Judging</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/https:/video.igem.org/w/aMwNT6eBryGs8xG5avXD51">Promotion Video</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/https:/igem.org/2021_Judging_Form?id=3711">Judging Form</a></div></li></ul></div></div></nav><div class="navbar-extra fixed-top"></div><header class="d-flex justify-content-center align-items-center"><div class="container"><h1 id="Top">Results</h1><p class="lead pl-1"></p><hr class="my-4"/></div></header><main><div class="container"><div class="row"><div class="sidebar col-lg-3"><div class="nav" id="contents"><h5>Contents</h5><ul></ul></div></div><div class="content col-lg-9"><article><h1>Introduction</h1><p>To realize hair dyeing & perming and decoloring & straightening as designed, we use E.coli to construct all the needed plasmid and transfect them into yeast through electroporation to express target enzymes and peptides. To confirm the validity of our perming and dyeing theory and eventually, the whole project, real hair is used in lab trails, which aim to estimate its color fastness, mechanical property, etc.</p><h1>Plasmid construction and amplification</h1><h2>The transformation of plasmid with AOX1 as promoter</h2><p>First of all, we need to amplificated all the commercially synthesized plasmid to acquire enough amount for further study. After transformation, colony PCR is applied for confirmation. Then we go for plasmid extraction.</p><div class="image"><img alt=" Colony PCR confirmation of successful E.coli transfection" src="https://static.igem.org/mediawiki/2021/8/8e/T--HUST-China--img--result--1.png" style="width: 100%"/><p>Figure 1: Colony PCR confirmation of successful E.coli transfection</p></div><p>Bright bands of identical sizes from colony PCR result demonstrates that target plasmid had successfully transformed into E.coli</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result1" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result1Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result1" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result1Label">result1</h5></div><div class="modal-body"><p>Plasmid with target gene is transformed into E.coli. From them, we acquire large amount of target gene using as raw material for further operation.</p><div class="image"><img alt="Colony PCR result of AOX1-&alpha; factor-ROX1-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/9/9b/T--HUST-China--img--result--2.png" style="width: 50%"/><p>Figure 2: Colony PCR result of AOX1-&alpha; factor-ROX1-AOX1 Terminator transformed E.coli</p></div><p>The band of AOX1-α factor-ROX1-AOX1 Terminator from colony PCR is about 3000bp, identical to the theoretical length of 3070bp estimated by the designed primer location (promoter to terminator), which could demonstrate that this target plasmid had successfully transformed into E.coli</p><div class="image"><img alt="Colony PCR result of AOX1-&alpha; factor-Laccase-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/5/50/T--HUST-China--img--result--3.png" style="width: 100%"/><p>Figure 3: Colony PCR result of AOX1-&alpha; factor-Laccase-AOX1 Terminator transformed E.coli</p></div><p>The band of AOX1-α factor-Laccase-AOX1 Terminator from colony PCR is about 3000bp, identical to the theoretical length of 3416bp estimated by the designed primer location (promoter to terminator), which could demonstrate that this target plasmid had successfully transformed into E.coli</p><div class="image"><img alt="Colony PCR results of AOX1-&alpha; factor-FMO-AOX1 Terminator, AOX1-&alpha; factor-crtE-AOX1 Terminator, AOX1-&alpha; factor-crtB-AOX1 Terminator and AOX1-&alpha; factor-crtI-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/5/55/T--HUST-China--img--result--4.png" style="width: 100%"/><p>Figure 4: Colony PCR results of AOX1-&alpha; factor-FMO-AOX1 Terminator, AOX1-&alpha; factor-crtE-AOX1 Terminator, AOX1-&alpha; factor-crtB-AOX1 Terminator and AOX1-&alpha; factor-crtI-AOX1 Terminator transformed E.coli</p></div><p>The bands of AOX1-α factor-FMO-AOX1 Terminator (3000+bp), AOX1-α factor-crtE-AOX1 Terminator (almost 3000bp), AOX1-α factor-crtB-AOX1 Terminator (less than 3000bp) and AOX1-α factor-crtI-AOX1 Terminator (3000+bp) from colony PCR are identical to the theoretical lengths of 3214bp, 2746bp, 2767bp and 3316 bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid had successfully transformed into E.coli</p><div class="image"><img alt="Colony PCR results of AOX1-&alpha; factor-CUS-AOX1 Terminator, AOX1-&alpha; factor-ACC-AOX1 Terminator, AOX1-&alpha; factor-4CL-AOX1 Terminator and AOX1-&alpha; factor-LOX2-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/4/46/T--HUST-China--img--result--5.png" style="width: 100%"/><p>Figure 5: Colony PCR results of AOX1-&alpha; factor-CUS-AOX1 Terminator, AOX1-&alpha; factor-ACC-AOX1 Terminator, AOX1-&alpha; factor-4CL-AOX1 Terminator and AOX1-&alpha; factor-LOX2-AOX1 Terminator transformed E.coli</p></div><p>The bands of AOX1-α factor-CUS-AOX1 Terminator (3000bp) , AOX1-α factor-ACC-AOX1 Terminator (3000+bp), AOX1-α factor-4CL-AOX1 Terminator (3000+bp) and AOX1-α factor-LOX2-AOX1 Terminator (almost 5000bp) from colony PCR are identical to the theoretical lengths of 3046bp, 3619bp, 3523bp and 4528bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid had successfully transformed into E.coli</p><div class="image"><img alt="Colony PCR results of AOX1-&alpha; factor-curA-AOX1 Terminator, AOX1-&alpha; factor-pepACS-AOX1 Terminator and AOX1-&alpha; factor-DsbC-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/2/2a/T--HUST-China--img--result--6.png" style="width: 100%"/><p>Figure 6: Colony PCR results of AOX1-&alpha; factor-curA-AOX1 Terminator, AOX1-&alpha; factor-pepACS-AOX1 Terminator and AOX1-&alpha; factor-DsbC-AOX1 Terminator transformed E.coli</p></div><p>The bands of AOX1-α factor-curA-AOX1 Terminator (almost 3000bp), AOX1-α factor-pepACS-AOX1 Terminator (almost 2000bp) and AOX1-α factor-DsbC-AOX1 Terminator (almost 3000bp) from colony PCR are identical to the theoretical lengths of 2875bp, 1987bp and 2722bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid had successfully transformed into E.coli</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h2>The construction of plasmid with Panb1 or Pynr071c as promoter</h2><p>AOX1 promoter is the strongest eukaryotic promoter currently known in yeast expression system. So we choose AOX1 as the primary promoter when we synthesized all these plasmid for the sake of more convenient expression. But noticing that methanol is hazardous, flammable, combustible and therefore, inappropriate to have direct contact with the hair, we need to substrate AOX1 for constitutive promoter Panb1 and xylose induced promoter Pynr071C to realize the projected regulation function as designed. Double-enzyme cleavage and rejointing is used to achieve this. We amplify the target gene located in the primarily synthesized plasmid without AOX1 promoter, and digest the acquired fragments and two kinds of plasmid, containing promoter only, with EcoR I and BamH I, then transform them into E.coli after linking the product together. Through this, we successfully substrate the primary AOX1 for Panb1 and Pynr071C.</p><div class="image"><img alt="Plasmid construction and colony PCR results of reconstructed plasmid with Panb1 and Pynr071C promoter" src="https://static.igem.org/mediawiki/2021/b/b7/T--HUST-China--img--result--7.png" style="width: 100%"/><p>Figure 7: Plasmid construction and colony PCR results of reconstructed plasmid with Panb1 and Pynr071C promoter</p></div><p>All the bands are identical to the theoretical lengths, which could demonstrate that these plasmid are correctly constructed and successfully transformed into E.coli, confirmed by sequencing.</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result2" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result2Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result2" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result2Label">result2</h5></div><div class="modal-body"><p>AOX1 promoter is the strongest eukaryotic promoter currently known in yeast expression system. So we choose AOX1 as the primary promoter when we synthesized all these plasmid for the sake of more convenient expression. But noticing that methanol is hazardous, flammable, combustible and therefore, inappropriate to have direct contact with the hair, we need to substrate AOX1 for constitutive promoter Panb1 and xylose induced promoter Pynr071C to realize the projected regulation function as designed. Another series of plasmid with target gene and new promoter are constructed.</p><div class="image"><img alt="Plasmid construction and colony PCR results of Panb1-&alpha; factor-FMO-AOX1 Terminator and Pynr071c-&alpha; factor-ROX1-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/d/d8/T--HUST-China--img--result--8.png" style="width: 100%"/><p>Figure 8: Plasmid construction and colony PCR results of Panb1-&alpha; factor-FMO-AOX1 Terminator and Pynr071c-&alpha; factor-ROX1-AOX1 Terminator transformed E.coli</p></div><p>The bands of Panb1-α factor-FMO-AOX1 Terminator (less than 3000bp) and Pynr071c-α factor-ROX1-AOX1 Terminator (3000+bp) from colony PCR are identical to the theoretical lengths of 2676bp and 3321bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p><div class="image"><img alt="Plasmid construction and colony PCR results of Panb1-&alpha; factor-4CL-AOX1 Terminator, Panb1-&alpha; factor-crtI-AOX1 Terminator, Panb1-&alpha; factor-crtB-AOX1 Terminator and Panb1-&alpha; factor-crtE-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/9/92/T--HUST-China--img--result--9.png" style="width: 100%"/><p>Figure 9: Plasmid construction and colony PCR results of Panb1-&alpha; factor-4CL-AOX1 Terminator, Panb1-&alpha; factor-crtI-AOX1 Terminator, Panb1-&alpha; factor-crtB-AOX1 Terminator and Panb1-&alpha; factor-crtE-AOX1 Terminator transformed E.coli</p></div><p>The bands of Panb1-α factor-4CL-AOX1 Terminator (3000+bp), Panb1-α factor-crtI-AOX1 Terminator (3000bp), Panb1-α factor-crtB-AOX1 Terminator (2000+bp) and Panb1-α factor-crtE-AOX1 Terminator (2000+bp) from colony PCR are identical to the theoretical lengths of 3185bp, 3046bp, 2198bp and 2177bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p><div class="image"><img alt="Plasmid construction and colony PCR results of Panb1-&alpha; factor-CUS-AOX1 Terminator, Panb1-&alpha; factor-ACC-AOX1 Terminator and Panb1-&alpha; factor-PepACS-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/6/6f/T--HUST-China--img--result--10.png" style="width: 100%"/><p>Figure 10: Plasmid construction and colony PCR results of Panb1-&alpha; factor-CUS-AOX1 Terminator, Panb1-&alpha; factor-ACC-AOX1 Terminator and Panb1-&alpha; factor-PepACS-AOX1 Terminator transformed E.coli</p></div><p>The bands of Panb1-α factor-CUS-AOX1 Terminator (2500bp), Panb1-α factor-ACC-AOX1 Terminator (3000bp) and Panb1-α factor-PepACS-AOX1 Terminator(less than 1500bp) from colony PCR are identical to the theoretical lengths of 2508bp, 3081bp and 1312bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h2>The construction of plasmid without signal peptide</h2><p>During the experiment, we find that not all the protein we want could be secreted extracellularly. Target protein is only detected in the yeast but not in the culture. while no signal appears in the supernatant without them, which indicates that not all of our signal peptides works. We reckon that as the α-factor on the plasmid we used is only a common basic signal peptide, which has some degree of universality but couldn't fit all the protein due to unseen matters like space structure of the protein, our target protein couldn't be induced to secret into the extracellular space by α-factor, or the low expression level coupled with degradation from protease existing in the extracellular environment could do the same. So, to get the rest of our enzymes which don't want to be outside of the cell with α-factor, and whether or not we could raise the expression level, we design new primers for PCR, which eliminate the signal peptide part off the whole target genes, and reconstruct the plasmid without the α-factor to verify if they prefer to be expressed inside of the cell.</p><div class="image"><img alt="Plasmid construction and colony PCR results of plasmid without signal peptides transformed E.coli" src="https://static.igem.org/mediawiki/2021/d/de/T--HUST-China--img--result--11.png" style="width: 100%"/><p>Figure 11: Plasmid construction and colony PCR results of plasmid without signal peptides transformed E.coli</p></div><p>All the bands are identical to the theoretical lengths, which could demonstrate that these plasmid are correctly constructed and successfully transformed into E.coli, confirmed by sequencing.</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result3" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result3Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result3" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result3Label">result3</h5></div><div class="modal-body"><p>Not quite to what we expect, after repeated transfection to the yeast, only a few products are expressed inside of eukaryotic system. Because of the large molecular weight and various types of some of our protein, we suspect that the common signal peptide we use, α-factor, is not enough to bring our protein out of the cell. While there is some of the genes without detectable products and we are hoping to get higher expression level, new primers for PCR are designed to ignore α-factor from our target gene in PCR. Then, likewise, we reconstruct this series of plasmid without α-factor through similar double-enzyme digestion and reconnection which insert our target genes right behind Panb1 promoter.</p><div class="image"><img alt="Plasmid construction and colony PCR results of Panb1-crtB-AOX1 Terminator, Panb1-crtE-AOX1 Terminator and Panb1-FMO-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/5/58/T--HUST-China--img--result--12.png" style="width: 100%"/><p>Figure 12: Plasmid construction and colony PCR results of Panb1-crtB-AOX1 Terminator, Panb1-crtE-AOX1 Terminator and Panb1-FMO-AOX1 Terminator transformed E.coli</p></div><p>The bands of Panb1-crtB-AOX1 Terminator (less than 2000bp), Panb1-crtE-AOX1 Terminator (less than 2000bp) and Panb1-FMO-AOX1 Terminator (2500bp) from colony PCR are identical to the theoretical lengths of 1859bp, 1838bp and 2437bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p><div class="image"><img alt="Plasmid construction and colony PCR results of Panb1-CUS-AOX1 Terminator, Panb1-ACC-AOX1 Terminator, Panb1-4CL-AOX1 Terminator and Panb1-crtI-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/8/8a/T--HUST-China--img--result--13.png" style="width: 100%"/><p>Figure 13: Plasmid construction and colony PCR results of Panb1-CUS-AOX1 Terminator, Panb1-ACC-AOX1 Terminator, Panb1-4CL-AOX1 Terminator and Panb1-crtI-AOX1 Terminator transformed E.coli</p></div><p>The bands of Panb1-CUS-AOX1 Terminator (2000+bp), Panb1-ACC-AOX1 Terminator (3000bp), Panb1-4CL-AOX1 Terminator (2500+bp) and Panb1-crtI-AOX1 Terminator (2500bp) from colony PCR are identical to the theoretical lengths of 2158bp, 2832bp, 2688bp and 2437bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h2>Exploring the construction of plasmid for pigment synthesis</h2><p>After all the expression product is detected, to explore the molecular mechanism laid behind and to save more experience for future teams, we tried to construct the complete whole pathways. Using seamless cloning, tentative experiments are performed by new primer for homologous recombination.</p><p>Unfortunately, no plasmid with multiple target genes are constructed. Good news is double-target-gene plasmid obtained. It seems that the identical promoters and terminators lead to the much shorter product of seamless cloning than theoretical, which means the enzymes used to join the designed homologous arms by primers falsely take some of the shared promoters and terminators as substrates as well. Although FMO dimers were successfully obtained, other plasmids were always shorter than expected .Then we give double-enzyme digestion and reconnection a try but limited to the restriction sites existed in our target genes and more fragments less odds and, well, time, no complete pathway is constructed. What we do have now is the precious experimental experience for all of our team members, and useful information for future teams.</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result4" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result4Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result4" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result4Label">result4</h5></div><div class="modal-body"><div class="image"><img alt="Plasmid construction and colony PCR result of Panb1-&alpha; factor -crtE-AOX1 Terminator-Panb1-&alpha; factor -crtI-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/4/40/T--HUST-China--img--result--14.png" style="width: 100%"/><p>Figure 14: Plasmid construction and colony PCR result of Panb1-&alpha; factor -crtE-AOX1 Terminator-Panb1-&alpha; factor -crtI-AOX1 Terminator transformed E.coli</p></div><p>The bands of Panb1-α factor-crtE-AOX1 Terminator-Panb1-α factor -crtI-AOX1 (less than 5000bp) and is identical to the theoretical lengths of 4574bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p><div class="image"><img alt="Plasmid construction and colony PCR result of Pynr071c-&alpha; factor-FMO dimer-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/0/0c/T--HUST-China--img--result--15.png" style="width: 100%"/><p>Figure 15: Plasmid construction and colony PCR result of Pynr071c-&alpha; factor-FMO dimer-AOX1 Terminator transformed E.coli</p></div><p>The bands of FMO dimer (less than 5000bp) and is identical to the theoretical lengths of 4740bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed. After serious of trails, it occurs that when multiple repeated fragments existed, seamless cloning may cause reduction of intended genes and of construction failure. We won't be able to finish the construction of complete pathway when going back for double-enzyme digestion and reconnection, which is much more difficult and complex.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Electroporation and expression of yeast</h1><p>To verify the expression of our genes and to acquire corresponding enzymes, we choose Pichia Pastoris GS115 as chassis and using electroporation to blend our plasmids into them. Large amount of target gene containing plasmid is extracted, then we digest them with Bgl II or Sal I to get linear plasmid, which could be integrated into yeast genome to avoid getting lost while being frozen. Concentration is also applied to achieve higher copy number and higher expression level.</p><p>After electroporation of plasmid with AOX1 as promoter and those without signal peptide, we acquire identical bands through Nickel-affinity chromatography column and SDS-PAGE.</p><p>For some of our enzymes don't have standard protocol to estimate their activity at present, we add substrates into culturing medium accordingly to find out whether there exists active target enzymes and do get our indigo and lycopene synthesized.</p><div class="image"><img alt="Medium for expression with substrates From left to right: GS115 medium with indole and FPP as control; Panb1-FMO-AOX1 Terminator medium with indole; mixture of Panb1-crtE-AOX1 Terminator, Panb1-crtB-AOX1 Terminator, Panb1-crtI-AOX1 Terminator medium with FPP" src="https://static.igem.org/mediawiki/2021/7/79/T--HUST-China--img--result--16.png" style="width: 100%"/><p>Figure 16: Medium for expression with substrates From left to right: GS115 medium with indole and FPP as control; Panb1-FMO-AOX1 Terminator medium with indole; mixture of Panb1-crtE-AOX1 Terminator, Panb1-crtB-AOX1 Terminator, Panb1-crtI-AOX1 Terminator medium with FPP</p></div><p>From left to right: GS115 medium with indole and FPP as control; Panb1-FMO-AOX1 Terminator medium with indole; mixture of Panb1-crtE-AOX1 Terminator, Panb1-crtB-AOX1 Terminator, Panb1-crtI-AOX1 Terminator medium with FPP</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result5" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result5Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result5" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result5Label">result5</h5></div><div class="modal-body"><p>Using E.coli for amplification, we extract and digest them with Bgl II or Sal I to get linear plasmid, which could be integrated into yeast genome to avoid getting lost while being frozen. Then, concentration of linear plasmid is also applied to achieve higher copy number and higher expression level. Several rounds of electroporation later, we successfully get all the plasmid with AOX1 as promoter into yeast.</p><div class="image"><img alt="Colony PCR result of yeast after electroporation" src="https://static.igem.org/mediawiki/2021/f/f3/T--HUST-China--img--result--17.png" style="width: 100%"/><p>Figure 17: Colony PCR result of yeast after electroporation</p></div><p>The bright bands are identical to the theoretical lengths, which could demonstrate that this target plasmid had successfully transformed into yeast.</p><p>After confirmation from colony PCR and sequencing, we using the successfully integrated yeast for expression. At first, we try to detect our target protein in the supernatant since there is signal peptide. At the same time, because the molecular weight of PepACS is only 5.08 kDa, it is almost impossible to separate from bromophenol blue, and its migration is also greatly affected, so the position of the band is not accurate. In order to judge whether it was expressed or not, we first filtered the miscellaneous proteins through 10kDa ultrafiltration tube, then concentrated it with 3kDa ultrafiltration tube, and then detected it by SDS-PAGE. Judge whether it is expressed successfully by whether there is coloring near the bottom line.</p><div class="image"><img alt="SDS-PAGE result of Laccase GS115 4CL LOX2 ACC pepACS DsbC+pepACS detecetion in the supernatant" src="https://static.igem.org/mediawiki/2021/9/9a/T--HUST-China--img--result--18.png" style="width: 100%"/><p>Figure 18: SDS-PAGE result of Laccase GS115 4CL LOX2 ACC pepACS DsbC+pepACS detecetion in the supernatant</p></div><p>Due to glycosylation modification of yeast expression, the molecular weight exhibited on SDS-PAGE will be larger than theoretical. Primary detection shows that we have laccase, 4CL and ACC bands of about 75kDa, LOX2 band of 100+kDa and DsbC+pepACS of about 40kDa, all of which is a bit larger(Laccase: 57.01 kDa; 4CL: 61.88 kDa; ACC: 63.40 kDa; LOX2: 102.88 kDa; DsbC+pepACS: 31.72 kDa) but still within explainable and acceptable range, which could be evidence of successful expression. There are irregular staining bands on the bromophenol blue line in the PepACS swimming lane. Although the molecular weight is not accurate, the successful expression can be judged according to the detection method we designed.</p><p>After confirmation of successful secret of some of the protein, we test the enzymatic activities of laccase and LOX2, which have standard activity testing protocol. Because of the thin band of laccase from SDS-PAGE, we can't be entirely sure whether it's inactive or active but too low the concentration to be detected. So, purification through Nickel-affinity chromatography column is used to raise its concentration for further test of enzymatic activity.</p><div class="image"><img alt="SDS-PAGE result of laccase after purification through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/c/c7/T--HUST-China--img--result--19.png" style="width: 100%"/><p>Figure 19: SDS-PAGE result of laccase after purification through Nickel-affinity chromatography column</p></div><p>Clear and thick band of about 75kDa is consistent to the result in the supernatant. Higher the eluent concentration, thicker the band. This indicates that the band of laccase in Fig18 is our target protein instead of other impurity with a high expression level.</p><p>During the test, we find a vague band of ROX1 which couldn't be verified as successfully expressed according to its SDS-PAGE result in the supernatant. As the key component of our xylose responding system, the successful expression of ROX1 is most vital to our project, so we apply purification through Nickel-affinity chromatography column to ROX1 to further identify its expression. Lucky us, target band is acquired and confirmed. It feels so good to announce that our ROX1 can be expressed like the others.</p><div class="image"><img alt="SDS-PAGE result of ROX1 after purification through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/2/21/T--HUST-China--img--result--20.png" style="width: 100%"/><p>Figure 20: SDS-PAGE result of ROX1 after purification through Nickel-affinity chromatography column</p></div><p>Different from impure or permeate bands, the target protein located around 55-55kDa, bigger than the theoretical 47.09kDa but still within explainable and acceptable range of glycosylation modification. ROX1 could be confirmed as successfully expressed. Although no gradient elution is applied, following elution result could verify this is ROX1, with some impurity bands occurring in the first ROX1 elution.</p><p>At the same time, after several SDS-PAGE confirmation, some of our proteins still don't want to show themselves. No band detected. We infer that this is due to A. the low level of extracellular expression and degradation by protease existing in the extracellular environment and B. the common signal peptide we use-α-factor-isn't specialized according to the unique space structure of our proteins so the mismatch causes failure of extracellular expression. To solve this, we reconstruct plasmids without the signal peptide and try to do intracellular expression. This is aim at all the undetectable or low-expressed genes.</p><div class="image"><img alt="Colony PCR result of yeast after electroporation of reconstructed plasmid without the signal peptide" src="https://static.igem.org/mediawiki/2021/d/db/T--HUST-China--img--result--21.png" style="width: 100%"/><p>Figure 21: Colony PCR result of yeast after electroporation of reconstructed plasmid without the signal peptide</p></div><p>The bright bands are identical to the theoretical lengths, which could demonstrate that this target plasmid had successfully transformed into yeast. Target genes are confirmed exist in the yeast of multiple bands, which could be the result of polluted electroporation cup.</p><p>After verification of successful transfection, we can't test the protein directly due to intracellular expression. So, we extract the total protein in yeast and go for a purification through Nickel-affinity chromatography column, then apply SDS-PAGE to separate target protein from the large amount and various type of total protein to confirm whether our target protein could be expressed and value its expression level quantitatively.</p><div class="image"><img alt="SDS-PAGE result of FMO after purification of yeast total protein extraction product through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/e/ed/T--HUST-China--img--result--22.png" style="width: 100%"/><p>Figure 22: SDS-PAGE result of FMO after purification of yeast total protein extraction product through Nickel-affinity chromatography column</p></div><p>Different from impure or permeate bands, the target protein located around 60kDa, bigger than the theoretical 53.96kDa but still within explainable and acceptable range of glycosylation modification. FMO could be confirmed as successfully expressed. The concentration of yeast total protein is so high that huge amount of impure protein is included during elution. But due to difference from impure or permeate bands, its dark color and consistency among several times of elution, this band could be verified as our target FMO,</p><div class="image"><img alt="SDS-PAGE result of crtE after purification of yeast total protein extraction product through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/1/16/T--HUST-China--img--result--23.png" style="width: 100%"/><p>Figure 23: SDS-PAGE result of crtE after purification of yeast total protein extraction product through Nickel-affinity chromatography column</p></div><p>Different from impure or permeate bands, the target protein located around 50kDa, bigger than the theoretical 33.42kDa but still within explainable and acceptable range of glycosylation modification. crtE could be confirmed as successfully expressed.</p><div class="image"><img alt="SDS-PAGE result of crtB after purification of yeast total protein extraction product through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/f/fc/T--HUST-China--img--result--24.png" style="width: 100%"/><p>Figure 24: SDS-PAGE result of crtB after purification of yeast total protein extraction product through Nickel-affinity chromatography column</p></div><p>Different from impure or permeate bands, the target protein located around 50kDa, bigger than the theoretical 35.30kDa but still within explainable and acceptable range of glycosylation modification. crtB could be confirmed as successfully expressed.</p><div class="image"><img alt="SDS-PAGE result of crtI after purification of yeast total protein extraction product through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/6/6f/T--HUST-China--img--result--25.png" style="width: 100%"/><p>Figure 25: SDS-PAGE result of crtI after purification of yeast total protein extraction product through Nickel-affinity chromatography column</p></div><p>Different from impure or permeate bands, the target protein located around 60kDa, bigger than the theoretical 55.86kDa but still within explainable and acceptable range of glycosylation modification. crtI could be confirmed as successfully expressed. After confirmation of successful expression, we add indole as substrate into culture medium of FMO and FPP into the mixture of crtE, crtB and crtI to test whether the expressed enzyme is active in terms of color change of the culture medium. As for the result, we succeed in expression of enzymes and synthesis of indigo and lycopene.</p><div class="image"><img alt="Medium for expression with substrates From left to right: GS115 medium with indole and FPP as control; Panb1-FMO-AOX1 Terminator medium with indole; mixture of Panb1-crtE-AOX1 Terminator, Panb1-crtB-AOX1 Terminator, Panb1-crtI-AOX1 Terminator medium with FPP" src="https://static.igem.org/mediawiki/2021/0/03/T--HUST-China--img--result--26.png" style="width: 100%"/><p>Figure 26: Medium for expression with substrates From left to right: GS115 medium with indole and FPP as control; Panb1-FMO-AOX1 Terminator medium with indole; mixture of Panb1-crtE-AOX1 Terminator, Panb1-crtB-AOX1 Terminator, Panb1-crtI-AOX1 Terminator medium with FPP</p></div><p>From left to right:</p><p>GS115 medium with indole and FPP as control; Panb1-FMO-AOX1 Terminator medium with indole; mixture of Panb1-crtE-AOX1 Terminator,Panb1-crtB-AOX1 Terminator,Panb1-crtI-AOX1 Terminator medium with FPP</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><p>Also, In order to verify whether the catalytic effect of FMO dimer is better than FMO. We electrotransformed the FMO dimer into yeast and tried to detect its expression and compare it with FMO.</p><p>The color of the medium and the cell were significantly darker than FMO under the same culture conditions. This shows that the catalytic activity of FMO dimer is stronger than that of FMO.</p><div class="image"><img alt="FMO dimer medium with indole after centrifugation.It is obvious that the culture medium and bacteria are dark blue." src="https://static.igem.org/mediawiki/2021/3/3a/T--HUST-China--img--result--27.png" style="width: 100%"/><p>Figure 27: FMO dimer medium with indole after centrifugation.It is obvious that the culture medium and bacteria are dark blue.</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result6" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result6Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result6" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result6Label">result6</h5></div><div class="modal-body"><p>After electrotransformed the FMO dimer into yeast, we still used Colony PCR to confirm the target gene is successfully transformed into the yeast cells.</p><div class="image"><img alt="Colony PCR result of yeast after electroporation of FMO dimer." src="https://static.igem.org/mediawiki/2021/e/ef/T--HUST-China--img--result--28.png" style="width: 100%"/><p>Figure 28: Colony PCR result of yeast after electroporation of FMO dimer.</p></div><p>The bands of FMO dimer (less than 5000bp) and is identical to the theoretical lengths of 4600bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that this target plasmid had successfully transformed into yeast.</p><p>At first, we tried to detect the protein in the supernatant, but no results were obtained. Because we have already experienced similar problems. We extracted the total protein directly and go for a purification to tested whether it was in the cell. Actually we detected the protein this time but it was smaller than expected.</p><div class="image"><img alt="SDS-PAGE result of FMO dimer after purification of yeast total protein extraction product through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/e/e6/T--HUST-China--img--result--29.png" style="width: 100%"/><p>Figure 29: SDS-PAGE result of FMO dimer after purification of yeast total protein extraction product through Nickel-affinity chromatography column</p></div><p>Different from impure or permeate bands, the target protein located around 60kDa, smaller than the theoretical 107.52kDa, but similar to the theoretical molecular weight of FMO (53.96kDa).</p><p>This indicated that the FMO dimer was broken in the process of expression or extraction. In order to confirm whether the catalytic effect is better, we also added indole to its medium to observe its effect on indigo synthesis.</p><div class="image"><img alt="FMOdimer medium with indole after centrifugation" src="https://static.igem.org/mediawiki/2021/e/ef/T--HUST-China--img--result--28.png" style="width: 100%"/><p>Figure 30: FMOdimer medium with indole after centrifugation</p></div><p>It is obvious that the culture medium and bacteria are dark blue.</p><p>The color of the medium and the cell were significantly darker than FMO under the same culture conditions. This shows that the catalytic activity of FMO dimer is stronger than that of FMO. So we can confirm that the expression is FMO dimer, and its effect is better. As for the problem of suspected fracture, we suspect that it is due to the short connection. It will withstand a large torsional strain when folding, resulting in its fragility, and fracture occurs in the process of extraction or sample preparation.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Determination of enzyme avtivity</h1><p>After the target strips appeared on SDS-PAGE, for the enzymes with standard enzyme activity assay such as Laccase and LOX2, we measured their enzyme activity according to the standard measurement methods to detect whether our enzymes are catalytically active. Laccase and LOX2 show high activity after adjusting the relevant conditions such as pH, temperature and ion concentration</p><div class="image"><img alt="Enzyme activity determination of Laccase" src="https://static.igem.org/mediawiki/2021/f/f2/T--HUST-China--img--result--31.png" style="width: 100%"/><p>Figure 31: Enzyme activity determination of Laccase</p></div><div class="image"><img alt="Enzyme activity determination of LOX2" src="https://static.igem.org/mediawiki/2021/0/02/T--HUST-China--img--result--32.png" style="width: 100%"/><p>Figure 32: Enzyme activity determination of LOX2</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result7" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result7Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result7" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result7Label">result7</h5></div><div class="modal-body"><p>After target strips been detected, to measure the activity of Laccase, we mixed 1ml of 1mmol / L ABTS solution with 1ml of centrifuged supernatant and adjusted the final concentration of Cu2+ to 10mmol/L, and measured its absorbance at 420nm. On this basis, we carried out determination experiments to further explore the optimal conditions for inducing the highest activity of Laccase.</p><p>Since the active center of Laccase is copper ion, we first explored the copper ion concentration to be added into the culture media. Adjusted the concentration of supplementary CuSO4 solution to 9mmol / L and 10mmol / L respectively, and measured the enzyme activity of Laccase under the same conditions</p><div class="image"><img alt="Effects of different concentrations of CuSO4 on enzyme activity" src="https://static.igem.org/mediawiki/2021/7/79/T--HUST-China--img--result--33.png" style="width: 100%"/><p>Figure 33: Effects of different concentrations of CuSO4 on enzyme activity</p></div><p>We found that the enzyme activity reaches its zenith when the concentration of copper ion is 10mmol/L. It is probably because the concentration of Cu2+ can't meet the need of Laccase active center when it is too low. When the concentration is too high, it will affect the binding of Laccase and substrate due to the existence of too much copper ions.</p><p>We also explored the effects of pH and temperature on activity of Laccase at the same time.</p><p>The optimum pH of Laccase was found to be acidic, therefore we prepared buffers with pH = 3, pH = 4.8 and pH = 6.6 respectively, with 100 mM citric acid and sodium citrate and added 1 ml buffer to control the pH of the reaction system while maintaining the final concentration of CuSO4 at 10 mmol / L</p><div class="image"><img alt="Effects of different pH on enzyme activity" src="https://static.igem.org/mediawiki/2021/a/a0/T--HUST-China--img--result--34.png" style="width: 100%"/><p>Figure 34: Effects of different pH on enzyme activity</p></div><p>After comparison, we found that among the three groups of data measured, Laccase activity is highest at pH = 3.0, decreasing to nadir at pH = 4.8, and completely inactivated at pH = 6.6. As for the reason why the subsequent measured value was lower than that at the beginning at pH 6.6, we speculated that it was caused by undermixing of solution when started reading.</p><p>For the effect of temperature, we kept ABTS solution and supernatant at 10℃, 20℃ and 37℃ respectively in advance, and kept the final concentration of CuSO4 at 10mmol / L.</p><div class="image"><img alt="" src="https://static.igem.org/mediawiki/2021/6/6a/T--HUST-China--img--result--35.png" style="width: 100%"/><p>Figure 35:</p></div><p>Through the analysis and comparison of the three groups of data, it is found that under testing conditions, the activity of Laccase is the highest when the temperature is 20℃, and the Laccase activity is inhibited by both low and high temperatures. As for the decrease of absorbance at the final phase of measuring enzyme activity at 10℃, it is suspected that the low temperature has a certain impact on ABTS cationic free radical, resulting in the decrease in absorbance</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Verification of promoters</h1><p>As we had successfully used Panb1 as promoter to express our protein, which can confirm that the Panb1 promoter can work smoothly. So to verify whether our Pynr071c promoter could initiate the expression of downstream genes, we linked GFP it, then transfect it into yeast and test the exist of fluorescence.</p><div class="image"><img alt="Fluorescence result of Pynr071c-GFP-AOX1 Terminator" src="https://static.igem.org/mediawiki/2021/d/d5/T--HUST-China--img--result--36.png" style="width: 100%"/><p>Figure 36: Fluorescence result of Pynr071c-GFP-AOX1 Terminator</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result8" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result8Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result8" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result8Label">result8</h5></div><div class="modal-body"><p>To verify whether our Pynr071c promoter could initiate the expression of downstream genes, we linked GFP it, then transfect it into yeast and test the exist of fluorescence.</p><div class="image"><img alt="Plasmid construction and colony PCR results of Pynr071c-GFP-AOX1 Terminator transformed E.coli" style="width: 100%"/><p>Figure 37: Plasmid construction and colony PCR results of Pynr071c-GFP-AOX1 Terminator transformed E.coli</p></div><p>The bands of Pynr071c-GFP-AOX1 Terminator from colony PCR are identical to the theoretical lengths of 2266bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p><p>We successfully construct the plasmid with the GFP and sequencing of which is correct. E.coli is used for amplification as well. Extract plasmid from E.coli, digest with Bgl II to get linear plasmid and concentrate for electroporation of yeast.</p><div class="image"><img alt="Colony PCR result of yeast after electroporation. The bright bands are identical to the theoretical lengths, which could demonstrate that this target plasmid had successfully transformed into yeast." src="https://static.igem.org/mediawiki/2021/3/34/T--HUST-China--img--result--38.png" style="width: 100%"/><p>Figure 38: Colony PCR result of yeast after electroporation. The bright bands are identical to the theoretical lengths, which could demonstrate that this target plasmid had successfully transformed into yeast.</p></div><p>We induce the plasmid with confirmation of colony PCR to express. Adding xylose everyday for xylose induced promoter Pynr071C to induce synthesis of GFP. Using fluorescence microscopy to detect the exist of green fluorescence, which indicates whether our promoters function regularly.</p><div class="image"><img alt="Fluorescence result of Pynr071c-GFP-AOX1 Terminator" src="https://static.igem.org/mediawiki/2021/4/42/T--HUST-China--img--result--39.png" style="width: 100%"/><p>Figure 39: Fluorescence result of Pynr071c-GFP-AOX1 Terminator</p></div></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>The Experiment Standard</h1><p>We measured the standard curves of three pigments before using them for hair dyeing experiment. We also found that the amount of melanin contained in hair can have a significant effect on hair dyeing outcomes. Therefore, we define different colors of hair based on bleaching.</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result1" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result1Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result1" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result1Label">result1</h5></div><div class="modal-body"><div class="image"><img alt="Curcumin concentration standard curve" src="https://static.igem.org/mediawiki/2021/8/89/T--HUST-China--img--result--result-curcumin_concentration_sc.png" style="width: 100%"/><p>Figure 40: Curcumin concentration standard curve</p></div><div class="image"><img alt="Indigo concentration standard curve" src="https://static.igem.org/mediawiki/2021/4/42/T--HUST-China--img--result--result-Indigo_concentration_sc.png" style="width: 100%"/><p>Figure 41: Indigo concentration standard curve</p></div><div class="image"><img alt="sLycopene concentration standard curve" src="https://static.igem.org/mediawiki/2021/5/55/T--HUST-China--img--result--result-Lycopene_concentration_sc.png" style="width: 100%"/><p>Figure 42: sLycopene concentration standard curve</p></div><div class="image"><img alt="Series of bleached hair" src="https://static.igem.org/mediawiki/2021/e/e9/T--HUST-China--img--result--result-the_sample_from_shop.png" style="width: 100%"/><p>Figure 43: Series of bleached hair</p></div><p>For the convenience of dyeing, we buy a series of bleached hair. The lightest color is the 9°hair, and the darkest is the 4 ° hair.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>The Best Condition of hair dye</h1><p>We have gained the best dye conditions of three kinds of hair dye(indigo, curcumin and lycopene) at a certain concentration. Under optimal conditions, we dyed 4-9 degrees of hair to get a series of dyeing discs. And we found that as for the three colors selected for the experiment, bleach the hair to 8 degrees could achieve a bright coloring effect.</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result2" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result2Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result2" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result2Label">result2</h5></div><div class="modal-body"><p>Chart of the best condition of hair dye</p><table><thead><tr><th>Dye/Condition</th><th>temperature</th><th>Dyeing aid ingredients</th><th>concentration(g/L)</th><th>comment</th></tr></thead><tbody><tr><td>indigo 2min</td><td>Room temperature</td><td>none</td><td>2</td><td>The color deepens significantly while dyeing for multiple times</td></tr><tr><td>curcumin 30min</td><td>50℃</td><td>Ethyl alcohol</td><td>0.5</td><td></td></tr><tr><td>lycopene 30min</td><td>Room temperature</td><td>alum</td><td>2</td><td></td></tr></tbody></table><p><em>The best dyeing condition</em></p><p>The exhibition of hair dyed on the best condition</p><p>Under the best conditions, we dyed the hair from 4 degree to 9 degree, and got a series of colors. It is found that it only needed to be bleached to 8 degree so that the hair would show a bright color for all three kinds of dye.</p><p>As to lycopene hair, 8 or 9 degree hair was red, 7 degree(or below) hair was brownish, and the longer the hair was dyed, the redder it would become.</p><p>As to indigo hair, 7 to 9 degree hair would become blue. As the dyeing time goes, the color would turn blue from an indigo color; 5 to 6 degree hair would be dyed to celadon, and 4 degree hair was still brown.</p><div class="image"><img alt="The dyeing results of lycopene(room temperature, adding alum, 2g/L). From left to right: 9&#176;(5,10,30min), 8&#176;(5,10,30min), 7&#176;(5,10,30min), 6&#176;(5,10,30min), 5&#176;(10,30min), 4&#176;(5,10,30min)" src="https://static.igem.org/mediawiki/2021/9/92/T--HUST-China--img--result--result-the_best_condition_for_lycopene_to_dye_.png" style="width: 100%"/><p>Figure 44: The dyeing results of lycopene(room temperature, adding alum, 2g/L). From left to right: 9&#176;(5,10,30min), 8&#176;(5,10,30min), 7&#176;(5,10,30min), 6&#176;(5,10,30min), 5&#176;(10,30min), 4&#176;(5,10,30min)</p></div><div class="image"><img alt="The dyeing results of indigo (room temperature, 2g/L). From left to right: 9&#176;(0.5,2,6min), 8&#176;(0.5,2,6min), 7&#176;(0.5,2,6min), 6&#176;(0.5,2,6min), 5&#176;(0.5,2,6min), 4&#176;(0.5,2,6min)" src="https://static.igem.org/mediawiki/2021/8/89/T--HUST-China--img--result--result-the_best_condition_for_indigo_to_dye.png" style="width: 100%"/><p>Figure 45: The dyeing results of indigo (room temperature, 2g/L). From left to right: 9&#176;(0.5,2,6min), 8&#176;(0.5,2,6min), 7&#176;(0.5,2,6min), 6&#176;(0.5,2,6min), 5&#176;(0.5,2,6min), 4&#176;(0.5,2,6min)</p></div><div class="image"><img alt="The dyeing results of indigo(room temperature, ethanol added, 0.5g/L). From left to right: 10min(9-4&#176;), 30min(9-4&#176;)8&#176;" src="https://static.igem.org/mediawiki/2021/e/e4/T--HUST-China--img--result--result-the_best_condition_for_turmeric_to_dye.png" style="width: 100%"/><p>Figure 46: The dyeing results of indigo(room temperature, ethanol added, 0.5g/L). From left to right: 10min(9-4&#176;), 30min(9-4&#176;)8&#176;</p></div><p>Problems occurred during the research:</p><p><strong>Lycopene</strong></p><p>Problem: No literature on coloring fabrics or hair with lycopene</p><p>Solution: We conducted a gradient experiment (0.5, 1, 2, 5 g/L) to explore the effective concentration of lycopene for hair dyeing. Finally, 2g/L of lycopene is selected, at which concentration the dye fluid will not be too viscous, and has a better dyeing effect as the picture below shows.</p><div class="image"><img alt="(From left to right: 0.5(30 min), 1(5, 10, 30 min), 2(5, 10, 30min), 5(5, 10, 30min)g/L of lycopene)" src="https://static.igem.org/mediawiki/2021/3/35/T--HUST-China--img--result--result-the_effort_to_confirm_the_best_concentration_for_lycopene_dying.png" style="width: 100%"/><p>Figure 47: (From left to right: 0.5(30 min), 1(5, 10, 30 min), 2(5, 10, 30min), 5(5, 10, 30min)g/L of lycopene)</p></div><p>Problem: Lycopene dye the hair with a low efficiency, a low color fastness, and a constantly discoloring process when the hair is showered by water.</p><p>Solution: We looked up the data and selected three eco-friendly color aids (alum, potassium tartrate, citric acid). Through direct color comparison and elution experiments, we found that alum can significantly improve the coloration rate and color fastness of lycopene.</p><div class="image"><img alt="(From left to right, 1st-4th groups: alum(30min), potassium tartrate(40min), citric acid(40 min), no color aids(40min); 5th-8th groups: the 1st-4th hair after washing 7 times)" src="https://static.igem.org/mediawiki/2021/6/6d/T--HUST-China--img--result--result-the_effort_for_confirming_the_maintaining_degree_of_lycopene.png" style="width: 100%"/><p>Figure 48: (From left to right, 1st-4th groups: alum(30min), potassium tartrate(40min), citric acid(40 min), no color aids(40min); 5th-8th groups: the 1st-4th hair after washing 7 times)</p></div><div class="image"><img alt="Color fastness test of lycopene" src="https://static.igem.org/mediawiki/2021/c/ca/T--HUST-China--img--result--result-Color_fastness_test_of_lycopene.png" style="width: 100%"/><p>Figure 49: Color fastness test of lycopene</p></div><p><strong>Curcumin</strong></p><p>Problem: The coloration rate of the curcumin aqueduct solution is low</p><p>Solution: We carried out the same dye addition and elution experiments as lycopene, and found that alum, potassium tartarate and citric acid cannot improve the coloring effect of curcumin. The data showed that curcumin is soluble in ethanol, and the optimal coloring temperature is 50 degree, so we adjusted the solvent to 33% ethanol solution. The process of coloring was conducted in the oven at 50 degrees, and the results showed that the coloring effect significantly improved.</p><div class="image"><img alt="Dyeing result of curcumin made in different solvents(30 min, 50&#176;C). Left: water solution, Right: 33% ethanol alcohol solution" src="https://static.igem.org/mediawiki/2021/c/c6/T--HUST-China--img--result--result-Dyeing_result_of_curcumin_made_in_different_solvents.png" style="width: 100%"/><p>Figure 50: Dyeing result of curcumin made in different solvents(30 min, 50&#176;C). Left: water solution, Right: 33% ethanol alcohol solution</p></div><p><strong>Indigo</strong> Problem: At the beginning of the experiment, we found that indigo could not be successfully adsorbed by the hair.</p><p>Solution: We looked up the data and found that in the printing and dyeing industry, it is necessary to restore the low solubility of indigo to water-soluble leucoindigo, which is used to color. We used the textile reducing agent - sodium sulfate to reduce indigo dye, and it came out to be successful.</p><p>Considering the safety of hair dyeing, we tried to use glucose as a reducing agent, by which we also obtained effective dyeing fluids. The method of using is indigo: glucose: caustic soda: 1:6:1, react for 15 min under 50°C. In the final product, we expected that yeast could conduct oxygen-free fermentation to reduce indigo with ethanol.</p><p>Problem: After dyeing your hair indigo, your hair becomes noticeably brittle.</p><p>Solution: Testing the average pull limit of a single hair, we found that the hair pull limit for indigo treatment was indeed significantly lower than that of normal hair. We assumed that this is related to the destruction of hair by alkaline substances. Therefore, we shorten the processing time of indigo dye, and finally designed to dye 2min at a time, repeated the procedure five times to achieve a saturation effect.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Hair stress experiment</h1><p>We used a micro scale experimental instrument to quantitatively evaluate the changes in mechanical properties of hair after hair dyeing. The results show that Indigo and lycopene as dyes have no obvious damage to hair. Curcumin has a repairing effect on hair.</p><div class="image"><img alt="Tensile Stress Strain Curves" src="https://static.igem.org/mediawiki/2021/e/ec/T--HUST-China--img--result--result-Tensile_Stress_Strain_Curves.png" style="width: 100%"/><p>Figure 51: Tensile Stress Strain Curves</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result3" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result3Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result3" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result3Label">result3</h5></div><div class="modal-body"><p>After hair dyeing, the mechanical properties of hair in each group changed to varying degrees. For example, the hair softens after a long treatment of the indigo solution becomes soft, elastic but easy to break. So we borrowed the micro scale experimental instrument independently developed by Professor Liu.</p><p>During the pre-experiment, we found there is no significant difference in ultimate stress between traditional dyeing hair and the untreated one, and we believe it's the added hair repair component causes the slug. Indigo and lycopene as dyes also have no obvious damage to hair. What's more, the yang's modulus of curcumin treatment hair is significantly higher. So we believe it keeps hair tough.</p><div class="image"><img alt="Indigo dyed hair being treated with micro tensile / micro torsional mechanical tester" src="https://static.igem.org/mediawiki/2021/b/b5/T--HUST-China--img--result--result-Indigo_dyed_hair_being_treated_with_micro_tensile_micro_torsional_mechanical_tester.png" style="width: 100%"/><p>Figure 52: Indigo dyed hair being treated with micro tensile / micro torsional mechanical tester</p></div></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Compound hair dye paste</h1><p>After finishing the solution experiment, we try to mix the natural pigment into a dye that can be applied directly to the hair. At present, lycopene dye and curcumin dye with NO.1 cream matrix as carrier are obtained, and natural essence is added to improve the odor of dye paste. Indigo is an oxidizing dye with special properties, so we designed a timely fermenter. In this way, we can use our product right now when indigo is produced and reduced to indigo white.</p><div class="image"><img alt="The expected product" src="https://static.igem.org/mediawiki/2021/5/55/T--HUST-China--img--result--result-the_expected_product.png" style="width: 100%"/><p>Figure 53: The expected product</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result4" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result4Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result4" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result4Label">result4</h5></div><div class="modal-body"><p><strong>Lycopene</strong></p><p>Difficulty: when we use lycopene paste to dye hair, it is not red but orange</p><p>Solution: by analyzing the cream formula, we think what is causing this problem is sodium sulfite. We add sodium sulfite to prevent further oxidation of the pigment, but it may also reduce and fade the pigment. The solution experiment proved our conjecture. A new lycopene dye that doesn't contain sodium sulfite found its way to red hair.</p><p><strong>Lycopene dying cream</strong></p><table><thead><tr><th>Ingredient</th><th>Content</th></tr></thead><tbody><tr><td>Cream matrix</td><td>100g</td></tr><tr><td>Sodium sulfite</td><td>0.2g</td></tr><tr><td>Absolute ethanol</td><td>1mL</td></tr><tr><td>pH 6.8 phosphate buffer</td><td>1mL</td></tr><tr><td>Solid paraffin</td><td>1 drop or not</td></tr><tr><td>Essence</td><td>1 drop</td></tr><tr><td>20% Lycopene</td><td>5g</td></tr><tr><td>Alum</td><td>0.6g</td></tr></tbody></table><p><strong>Curcumin</strong> EDTA is added to the curcumin paste to prevent metal ions from interfering with the effect -- for example, Fe3 + producing a reddish-brown tint.</p><p><strong>Curcumin dying cream</strong></p><table><thead><tr><th>Ingredient</th><th>Content</th></tr></thead><tbody><tr><td>Cream matrix</td><td>100g</td></tr><tr><td>EDTA</td><td>0.2g</td></tr><tr><td>Sodium sulfite</td><td>0.1g</td></tr><tr><td>Absolute ethanol</td><td>1mL</td></tr><tr><td>pH 6.8 phosphate buffer</td><td>1mL</td></tr><tr><td>Isopropyl alcohol</td><td>2mL</td></tr><tr><td>Solid paraffin</td><td>1 drop or not</td></tr><tr><td>Essence</td><td>1 drop</td></tr><tr><td>curcumin</td><td>0.5g</td></tr></tbody></table><div class="image"><img alt="The result of curcumin staining cream. 9 &#176; from left to right. Hair was dyed at room temperature for 30min, 60min and 0min; Dyeing at 50 &#8451; for 10min and 30min" src="https://static.igem.org/mediawiki/2021/a/a0/T--HUST-China--img--result--result-the_different_effort_of_termeric_dying_at_changing_tep.%26time.png" style="width: 100%"/><p>Figure 54: The result of curcumin staining cream. 9 &#176; from left to right. Hair was dyed at room temperature for 30min, 60min and 0min; Dyeing at 50 &#8451; for 10min and 30min</p></div><p>The result of curcumin staining cream. 9 ° from left to right. Hair was dyed at room temperature for 30min, 60min and 0min; Dyeing at 50 ℃ for 10min and 30min</p><p><strong>Indigo:</strong></p><p>Difficult: we can make indigo paste, but the hair does not dye well.</p><p>Solution: Indigo is a water-soluble component, and need to be oxidized to indigo after fixing to the hair. With water-in-oil paste as the matrix, indigo white can not fully enter the interior of the hair, and the oily substances in the matrix and excessive reductant prevent indigo white from oxidation in the hair, resulting in no effective coloring.</p><p>So we decided to design a timely manner in which indigo could be produced and used at the same time. Therefore, we consider that indigo dye can be produced and used in time -- the direct production of indigo by yeast, and the production of indigo solution as a dye in time.</p><p>For this idea, we dye indigo solution directly on the hair and find that it can be painted, but it can not color the hair evenly. So we designed a hair dye comb to make it possible to evenly smear indigo cryptosomes on the hair. The matching device is a timely fermentation tank, which can meet the needs of users with our engineering bacteria as raw materials, timely production and timely use of indigo white. For detailed information, please refer to <a href="https://2021.igem.org/Team:HUST-China/Results/Hardware">Hardware</a> part.</p><div class="image"><img alt="the different effort of indigo" src="https://static.igem.org/mediawiki/2021/5/5b/T--HUST-China--img--result--result-the_different_effort_of_indigo_dying_at_changing_tep.%26time.png" style="width: 100%"/><p>Figure 55: the different effort of indigo</p></div></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Color fastness test</h1><p>Color fastness is an important aspect to measure the effect of dye, so we design a set of elution scheme and test the color fastness of three kinds of natural pigment dye products and the same color traditional dye paste. The results showed that the color fastness of the natural pigment dyes was better than that of the traditional dyes.</p><div class="image"><img alt="The comparison of biological dying with traditional dying" src="https://static.igem.org/mediawiki/2021/e/ee/T--HUST-China--img--result--result-the_comparison_of_biological_dying_with_traditional_dying.png" style="width: 100%"/><p>Figure 56: The comparison of biological dying with traditional dying</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result5" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result5Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result5" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result5Label">result5</h5></div><div class="modal-body"><p>The experiment of color fastness:</p><p>Compound 25g/L shampoo solution. Add 10 ML shampoo solution and a bunch of hair in a 15 ML centrifuge tube for 5 min at 100 r/min. The experiment was repeated seven times. The absorbance of the eluent at the maximum absorption band of the corresponding color was measured and the curve was drawn. Results as shown in figure, the single elution amount of three kinds of natural pigment was less than that of the same color traditional dyes, and the color change after 7 times elution was also less than that of the traditional dyes. Furthermore, curcumin dyeing cream treatment of hair is almost non-decolorization.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Toxicological experiment of broad bean root tip</h1><p>In order to test the safety of our dye paste, we carried out a toxicology experiment of broad bean root tip. The rate of micronucleus in root tip cells on broad bean was measured. The higher the micronucleus rate was, the higher the teratogenic rate was. The results showed that the teratogenicity of our product was significantly lower than that of the two traditional hair dyes on the market, indicating that our product was less toxic.</p><div class="image"><img alt="Micronucleus Frequency" src="https://static.igem.org/mediawiki/2021/0/0a/T--HUST-China--img--result--result-micronucleus_frequency.png" style="width: 100%"/><p>Figure 57: Micronucleus Frequency</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result6" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result6Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result6" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result6Label">result6</h5></div><div class="modal-body"><p>Micronucleus (MCN) is an abnormal structure in eukaryotic cells, which is often produced by the action of radiation or chemical drugs. In the intercellular phase, the Micronuclei were round or oval, free from the main nucleus, the size should be less than 1/3 of the main nucleus. Only the eucaryotic test system can directly predict the genetic damage of mutagens to human beings and other higher organisms. In this respect, the micronucleus test is an ideal method. And some studies show that the plant micronucleus test and the animal test have a coincidence rate of up to 99%. Micronucleus test using broad bean root tip as experimental material can accurately show the effect of various treatments on induced distortion.</p><div class="image"><img alt="Patricles" src="https://static.igem.org/mediawiki/2021/d/d7/T--HUST-China--img--result--result-patricles.png" style="width: 100%"/><p>Figure 58: Patricles</p></div><p>Specific experimental process:</p><ol><li>We bought 2 catties of sun-dried broad bean seeds, put them in a wet porcelain tray after soaking in water overnight. After 2 days, about 95% of the bean seeds grew roots about 1-2 cm long.</li><li>We divided 10 broad beans into 1 group. Each group of root tips were immersed in the following concentrations of dye paste solution for 24 hours.</li><li>We took the broad beans soaked in the dye solution, washed them with distilled water, then put them into distilled water and cultured for 24 hours.</li><li>The root tips of Vicia Faba were cut off and fixed in 15-fold Kano Solution (anhydrous ethanol: Glacial Acetic acid = 3:1) for 24 hours.</li><li>We took out the root tip, washed it with distilled water, put it into 1 m hydrochloric acid solution, and reacted at 60 °C for 20 minutes to soften the root tip. The root tip was cut 1 mm, smashed on the slide, stained with the modified carbolic acid red dye for 15 min, covered with the slide, then flattened and observed under the microscope.</li><li>We first find the meristematic cells, which are closely arranged and square, look at 1000 cells, and record the number of cells with micronucleus. Micronucleus rate = micronucleus cell number/1000 ?? 1000%</li></ol><table><thead><tr><th>Indigo</th><th>Traditional blue</th><th>Lycopene</th><th>Traditional red</th><th>curcumin</th><th>Negative control:Distilled water</th><th>Positive control:1g/L CuSO4</th></tr></thead><tbody><tr><td>9g/L</td><td>9g/L</td><td>9g/L</td><td>9g/L</td><td>9g/L</td><td></td><td></td></tr><tr><td>6g/L</td><td>6g/L</td><td>6g/L</td><td>6g/L</td><td>6g/L</td><td></td><td></td></tr><tr><td>4.5g/L</td><td>4.5g/L</td><td>4.5g/L</td><td>4.5g/L</td><td>4.5g/L</td><td></td><td></td></tr><tr><td>3g/L</td><td>3g/L</td><td>3g/L</td><td>3g/L</td><td>3g/L</td><td></td><td></td></tr><tr><td>1.5g/L</td><td>1.5g/L</td><td>1.5g/L</td><td>1.5g/L</td><td>1.5g/L</td><td></td><td></td></tr></tbody></table><div class="image"><img alt="Broad bean dyed with pigment" src="https://static.igem.org/mediawiki/2021/7/70/T--HUST-China--img--result--result-broad_bean_dyed_with_pigment.png" style="width: 100%"/><p>Figure 59: Broad bean dyed with pigment</p></div></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><p>Due to the huge workload of counting micronucleus, we tried to develop counting algorithm. We use the algorithm framework of Yolov5 to implement what is available on the author's Github or Model section, which is not covered here. We selected the typical normal nuclei and Micronuclei from the frame of the microscope. There were about 200 chromatin images and 50 micronuclei images in the normal nuclei. We trained a recognition model by using these images as training set. With this model, a large number of imaging results can be analyzed in a short time. We use this model to count micronuclei and measure the cytotoxicity of various substances. The results show that this model is in good agreement with the commonly used counting standards under the experimental conditions. It can be used to compare the difference in toxicity between different substances. For detailed information, please refer to <a href="https://2021.igem.org/Team:HUST-China/Results/Model">Model</a> part. #Color stability testing We would expose our hair dyed with our dye paste and two chemical dyes (red and blue) to constant bright light, take pictures before and after lighting, and calculate the degree of discoloration. The result shows that the light stability of our dye paste is slightly less than that of the chemical dye paste, but the difference is invisible to the naked eye in the actual application scene, so the color of our dye paste can exist stably in the natural light, it's still very practical.</p><div class="image"><img alt="Light exposure's influence on dying" src="https://static.igem.org/mediawiki/2021/f/ff/T--HUST-China--img--result--result-light_exposure%27s_influence_on_dying_.png" style="width: 100%"/><p>Figure 60: Light exposure's influence on dying</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result7" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result7Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result7" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result7Label">result7</h5></div><div class="modal-body"><p>What we did: We cut a bunch of hair in half, pasted half of it onto a piece of cardboard, and exposed it to a constant, bright light for 11 days. Our lights are LED lights and sodium lamps, cycle for 22 hours of light and 2 hours of darkness alternately. At the end of the illumination, half of the hair without illumination was pasted on, and the fade index was analyzed by the algorithm. Considering the change of RGB values, it is difficult to determine the model parameters, and there may be multi-value based on the prediction results given by the neural network. We choose the linear combination of RGB to calculate the gray value, and take the gray value as the index of the concentration. It can be used as an index to evaluate the degree of fading by calculating the change of gray difference between the two groups. This indicator is not linear and therefore should not be understood as an indicator in a percentage sense. On a scale of one to one, the higher the score, the deeper the discoloration. There was no significant difference between indigo and chemical blue, the scores of indigo were 0.178 and 0.161, respectively. Chemical red faded more slowly than lycopene, with a score of 0.186 and 0.313, respectively.</p><p>The process of exploration: When we first carried out the strong light experiment, we irradiated single hair and only treated it for 2 days. The results showed that the dyeing of each hair in the same strand was very different due to the uneven dyeing, and the treatment time is so short that almost all the hair remains the same. In the second experiment, we took a photo of the hair in front of the light and compared it with the photo after the light exposure. The light exposure was extended to 7 days. Although the discoloration was detectable this time, it varied widely between the groups, suggesting that one hair's discoloration was highly accidental. So on the third occasion, we decided to reduce the chance by increasing the amount of hair, using a single strand of hair, and extending the light again to get the difference.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Color matching experiment and Sumu experiment</h1><p>By adding color aids, mixing two dyes or secondary coloring, we get more hair colors. Now, we have different brightness of the rainbow seven colors and gray, red, brown, tawny and other colors. At the same time, we also used hematoxylin as a dye to explore a series of dyeing. Hematoxylin itself can be brown, add alum can be red, add alkali can be purple. Hematoxylin 's synthetic biology production will be a future extension project.</p><div class="image"><img alt="Olympic rings and Cartoon kid" src="https://static.igem.org/mediawiki/2021/0/04/T--HUST-China--img--result--result-Olympic_rings_and_Cartoon_kid.png" style="width: 100%"/><p>Figure 61: Olympic rings and Cartoon kid</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result8" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result8Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result8" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result8Label">result8</h5></div><div class="modal-body"><p>A coloring list of the three pigments</p><table><thead><tr><th>Picture</th><th>Color</th><th>Coloring method</th></tr></thead><tbody><tr><td><img src="https://static.igem.org/mediawiki/2021/d/d9/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_red.png"/></td><td>Red</td><td>8 ° hair in the room temperature treated with lycopene for 30 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/e/e9/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_orange.png"/></td><td>Orange</td><td>9 ° hair in the room temperature treated with curcumin for 30 minutes then treated with lycopene for another 30 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/6/67/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_light_orange.png"/></td><td>Light orange</td><td>9 ° hair in the room temperature treated with curcumin mixed with lycopene (2:1) for 30 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/1/17/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_yellow.png"/></td><td>Yellow</td><td>9 ° hair in 50 ℃ treated with curcumin for 30 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/f/fc/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_green.png"/></td><td>Green</td><td>7 ° hair in the room temperature treated with indigo for 2 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/2/29/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_cyan.png"/></td><td>Cyan</td><td>9 ° hair in the room temperature treated with indigo for 30 seconds</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/8/89/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_blue.png"/></td><td>Blue</td><td>9 ° hair in the room temperature treated with indigo for 2 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/7/78/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_purple.png"/></td><td>Purple</td><td>9 ° hair in the room temperature treated with indigo for 5 minutes then treated with lycopene for another 10 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/0/0b/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_grey.png"/></td><td>Gray</td><td>9 ° hair in the room temperature treated with lycopene for 5 minutes then treated with indigo for another 30 seconds</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/5/5b/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_reddish_brown.png"/></td><td>Reddish brown</td><td>6 ° hair in the room temperature treated with lycopene for 10 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/8/8b/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_tan.png"/></td><td>Tan</td><td>9 ° hair in the room temperature added tannic acid treated with curcumin for 10 minutes</td></tr></tbody></table></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><div class="image"><img alt="the coloring effect of hematoxylin under various conditions" src="https://static.igem.org/mediawiki/2021/7/7b/T--HUST-China--img--result--result-the_overview_of_all_possible_colors.png" style="width: 100%"/><p>Figure 62: the coloring effect of hematoxylin under various conditions</p></div><h1>Experiment of short peptide perm</h1><h2>Short peptides perming hair experiment</h2><div class="image"><img alt="comparison of short peptides and water" src="https://static.igem.org/mediawiki/2021/6/61/T--HUST-China--img--result--result-comparison_of_short_peptides_and_water.png" style="width: 100%"/><p>Figure 63: comparison of short peptides and water</p></div><p>We used 0.1% and 0.01% short peptides for perm experiment, and set chemical perm and up water perm as control group. The perming effect of short peptide is more obvious than that of ultra-pure water, which shows that in alkaline and 50° conditions, the hair cuticle is opened and the longer react time it takes, the better perming result we get. And after a series experiment, we found that hair dealt with SDS can have more obvious curl effect.</p><div class="image"><img alt="comparison of short peptides with different mixture with other chemicals" src="https://static.igem.org/mediawiki/2021/7/78/T--HUST-China--img--result--result-comparison_of_short_peptides_with_different_mixture_with_other_chemicals.png" style="width: 100%"/><p>Figure 64: comparison of short peptides with different mixture with other chemicals</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result9" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result9Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result9" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result9Label">result9</h5></div><div class="modal-body"><table><thead><tr><th style="text-align:center">Chemical perm agent(bought online)</th><th>Ultra-pure water</th><th>0.001% short peptide solution</th><th>0.1% short peptide solution</th></tr></thead><tbody><tr><td colspan="4" style="text-align:center">500ul</td></tr><tr><td colspan="4" style="text-align:center">Immerse hair(washed and dried) 20min</td></tr></tbody></table><p>The experimental results showed that the Chemical perm agent had the most obvious effect, while others didn't have any obvious perming effect. We've considered 3 reasons why short peptides have no obvious effect:</p><p>(1) Maybe the peptides didn't get into hair cuticle</p><p>(2) The time for short peptides to form disulfide bonds wasn't enough</p><p>(3) The reaction temperature is not enough.</p><p>So, we get another exploring experiment.</p><p>Time and Temperature:</p><p>(1)Drop a drop of 3M NaOH into 0.01% short peptides solution. [PH test paper: PH=10]</p><p>(2)Control experiment: drop a drop of 3M NaOH into UP water</p><p>(3)Both react at 50° for 1h.</p><p>Re-explore a better condition for short peptides'perming hair</p><p>1)immerse 10 hairs in SDS 3 times to remove charges on hair surface.</p><p>2)Wash and put them in 0.01% short peptides solution</p><p>3)Control experiment: Up water, the subsequent processing steps are the same as 1)2)</p><p>4)both:50°, 1h</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Perming hair stress experiment-9</h1><p>We used different kinds of perms for hairs with 2 groups of different hair, and the hair strains will be damaged after perming. So we designed and conducted stress experiments to test the mechanics of the hair after perming. The figure below shows the experiment data, by which we can know that perming by short peptides will have very little damage to the hair, and short peptide perm damage is less than chemical perm. Moreover, the results of the damaged hair groups showed that the damage of short peptide perm is much smaller than that of chemical perm. Hence, we speculated that perming procedure would cause some damage to hair, but the use of short peptide perm could be used to repair the damaged hair.</p><h3>Healthy hair stress test results (Horizontal: the deformation length; Ordinate: pull force</h3><div class="image"><img alt="Healthy hair stress test results (Horizontal: the deformation length; Ordinate: pull force)" src="https://static.igem.org/mediawiki/2021/4/4b/T--HUST-China--img--result--result-healthy_hair_stress_test_result.png" style="width: 100%"/><p>Figure 65: Healthy hair stress test results (Horizontal: the deformation length; Ordinate: pull force)</p></div><p>It is shown that the hair of the short peptide perm group changes in shape under constantly increasing pull force, and most hair strains can withstand more pull than hair permed by chemical perms. This suggests that short peptide perms do less damage to hair performance than chemical perms. According to the experimental results, we judge that the concentration of short peptides has little effect on the results.</p><h3>Damaged hair stress test results (Horizontal: the maximum length; Ordinate: pull force)</h3><div class="image"><img alt="Damaged hair stress test results (Horizontal: the maximum length; Ordinate: pull force" src="https://static.igem.org/mediawiki/2021/e/e9/T--HUST-China--img--result--result-damaged_hair_stress_test_result.png" style="width: 100%"/><p>Figure 66: Damaged hair stress test results (Horizontal: the maximum length; Ordinate: pull force</p></div><p>This set of data comes from a pulling test of damaged hair. It is shown in this figure that both sets of hairs that use short peptides for perm can withstand more pull than the hairs after chemical perm. And there isn't much difference between the results of short peptide group and that of the water group. We speculate that short peptides can by some means repair the damaged hair, which also portrays the excellent performance of short peptide perm that it can protect the hair and maintain the hair quality.</p><h1>Microscopic effects of PepACS on hair</h1><p>We used scanning electron microscopy to look at healthy hair, bleached hair and bleached hair treated with the PepACS. The results showed that compared with normal hair, the surface of bleached hair was obviously damaged, but no obvious damage was observed after treatment with PepACS, indicating that PepACS may have a certain repair effect, which can be further studied in the future.</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result10" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result10Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result10" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result10Label">result10</h5></div><div class="modal-body"><p>We soaked the damaged hair with 0.1% PepACS solution and reacted at 50℃ for 3 hours to obtain the damaged hair samples treated with PepACS. No surface damage was clearly observed under scanning electron microscopy after the PepACS treatment, but given the differences between the same batch of damaged hair, we cannot say that the PepACS has a repairing effect. We expect to continue to carry out relevant experiments in the future.</p><div class="image"><img alt="From left to right: healthy hair,bleached hair,bleached hair treated with the PepACS" src="https://static.igem.org/mediawiki/2021/4/4b/T--HUST-China--img--result--result-elect.png" style="width: 100%"/><p>Figure 67: From left to right: healthy hair,bleached hair,bleached hair treated with the PepACS</p></div></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><div class="row g-0"><div class="col-4 girl"><img src="https://static.igem.org/mediawiki/2021/2/28/T--HUST-China--img--girl.png"/></div><div class="col-5"><div class="dialogbox"><p>Wow! That’s eye-opening! I guess the old saying that “Rome was not build in a day” does make sense. All those results require accumulation, which is recorded in the <a href="https://2021.igem.org/Team:HUST-China/Results/Results">notebook</a>.</p></div></div></div></article></div></div></div></main><footer><div class="container"><div class="row"><div class="col-7"><div class="row justify-content-center"><div class="footer-logo"></div></div><p><span>CONTACT US: </span><a href="mailto:iGEMHUSTChina@163.com">iGEMHUSTChina@163.com</a></p><p>Huazhong University of Sci. & Tech., Wuhan, China</p><p>1037# Luoyu Rd, Wuhan, P.R.China 430074</p><p>Copyright © <span>HUST-China </span>iGEM 2021</p></div><div class="col-5"><div class="row"><div class="footer-xmind"></div></div><div class="row"><div class="footer-snapgene"></div></div><div class="row"><div class="footer-NEBI"></div></div></div></div></div></footer><script src="https://2021.igem.org/Template:HUST-China/content-bundleJS?action=raw&ctype=text/javascript"></script><script src="https://2021.igem.org/Template:HUST-China/mathjax-bundleJS?action=raw&ctype=text/javascript"></script></body></html>
| + | <!-- # TODO: #6 Fix table caption font--><!-- # TODO: #7 Fix citations links font size--><html lang="en"><head><meta charset="utf-8"/><meta content="width=device-width,initial-scale=1" name="viewport"/><link href="https://2021.igem.org/Template:HUST-China/css/contentCSS?action=raw&ctype=text/css" rel="stylesheet"/><title>Results | iGEM HUST-China</title><script>MathJax={tex:{inlineMath: [['$', '$'], ['\(', '\)']],packages: {'[+]': ['mhchem']},loader: {load: ['[tex]/mhchem']},};</script><link href="https://2021.igem.org/Template:HUST-China/css/contentCSS?action=raw&ctype=text/css" rel="stylesheet"/></head><body><!-- # TODO: #6 Fix table caption font--><!-- # TODO: #7 Fix citations links font size--><nav class="navbar navbar-expand-xl fixed-top"><div class="container d-flex justify-content-between"><a class="navbar-brand" href="https://2021.igem.org/Team:HUST-China"><i class="navbar-logo-left"></i><span>HUST-China</span></a><button aria-controls="navbarNav" aria-expanded="false" aria-label="Toggle navigation" class="navbar-toggler" data-target="#navbarNav" data-toggle="collapse" type="button"><span class="navbar-toggler-icon"></span></button><div class="collapse navbar-collapse" id="navbarNav"><ul class="navbar-nav ml-auto"><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarProjectDropdown" role="button">Project</a><div aria-labelledby="navbarProjectDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Description">Description</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Design">Design</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Proof_Of_Concept">Proof of Concept</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Safety">Safety</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Implementation">Proposed Implementation</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Contribution">Contribution</a></div></li><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarHuman PracticesDropdown" role="button">Human Practices</a><div aria-labelledby="navbarHuman PracticesDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Human_Practices">Overview</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Human_Practices_01">Part 1</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Human_Practices_02">Part 2</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Collaborations">Part 3</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Education">Part 4</a></div></li><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarModelingDropdown" role="button">Modeling</a><div aria-labelledby="navbarModelingDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Model">Overview</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Gene_Pathway">Gene Pathway</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Dye_color_forecast">Dye color forecast</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Micronucleus_Counter">Micronucleus Counter</a></div></li><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarProductsDropdown" role="button">Products</a><div aria-labelledby="navbarProductsDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Products">Overview</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Software">Software</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Hardware">Hardware</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Another_Area">Another Area</a></div></li><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarExperimentsDropdown" role="button">Experiments</a><div aria-labelledby="navbarExperimentsDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Experiments">Overview</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Parts">Parts</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Engineering">Engineering Success</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Results">Results</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Protocol">Protocol</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Notebook">Notebook</a></div></li><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarPeopleDropdown" role="button">People</a><div aria-labelledby="navbarPeopleDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Team">Team</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Partnership">Partnership</a><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Attributions">Attributions</a></div></li><li class="nav-item dropdown"><a aria-expanded="false" aria-haspopup="true" class="nav-link dropdown-toggle" data-toggle="dropdown" href="#" id="navbarAchievementsDropdown" role="button">Achievements</a><div aria-labelledby="navbarAchievementsDropdown" class="dropdown-menu"><a class="dropdown-item" href="https://2021.igem.org/Team:HUST-China/Judging">Judging</a><a class="dropdown-item" href="https://video.igem.org/w/aMwNT6eBryGs8xG5avXD51">Promotion Video</a><a class="dropdown-item" href="https://igem.org/2021_Judging_Form?id=3711">Judging Form</a></div></li></ul></div></div></nav><div class="navbar-extra fixed-top"></div><header class="d-flex justify-content-center align-items-center"><div class="container"><h1 id="Top">Results</h1><p class="lead pl-1"></p><hr class="my-4"/></div></header><main><div class="container"><div class="row"><div class="sidebar col-lg-3"><div class="nav" id="contents"><h5>Contents</h5><ul></ul></div></div><div class="content col-lg-9"><article><h1>Introduction</h1><p>To realize hair dyeing & perming and decoloring & straightening as designed, we use E.coli to construct all the needed plasmid and transfect them into yeast through electroporation to express target enzymes and peptides. To confirm the validity of our perming and dyeing theory and eventually, the whole project, real hair is used in lab trails, which aim to estimate its color fastness, mechanical property, etc.</p><h1>Plasmid construction and amplification</h1><h2>The transformation of plasmid with AOX1 as promoter</h2><p>First of all, we need to amplificated all the commercially synthesized plasmid to acquire enough amount for further study. After transformation, colony PCR is applied for confirmation. Then we go for plasmid extraction.</p><div class="image"><img alt=" Colony PCR confirmation of successful E.coli transfection" src="https://static.igem.org/mediawiki/2021/8/8e/T--HUST-China--img--result--1.png" style="width: 100%"/><p>Figure 1: Colony PCR confirmation of successful E.coli transfection</p></div><p>Bright bands of identical sizes from colony PCR result demonstrates that target plasmid had successfully transformed into E.coli</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result1" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result1Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result1" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result1Label">result1</h5></div><div class="modal-body"><p>Plasmid with target gene is transformed into E.coli. From them, we acquire large amount of target gene using as raw material for further operation.</p><div class="image"><img alt="Colony PCR result of AOX1-&alpha; factor-ROX1-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/9/9b/T--HUST-China--img--result--2.png" style="width: 50%"/><p>Figure 2: Colony PCR result of AOX1-&alpha; factor-ROX1-AOX1 Terminator transformed E.coli</p></div><p>The band of AOX1-α factor-ROX1-AOX1 Terminator from colony PCR is about 3000bp, identical to the theoretical length of 3070bp estimated by the designed primer location (promoter to terminator), which could demonstrate that this target plasmid had successfully transformed into E.coli</p><div class="image"><img alt="Colony PCR result of AOX1-&alpha; factor-Laccase-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/5/50/T--HUST-China--img--result--3.png" style="width: 100%"/><p>Figure 3: Colony PCR result of AOX1-&alpha; factor-Laccase-AOX1 Terminator transformed E.coli</p></div><p>The band of AOX1-α factor-Laccase-AOX1 Terminator from colony PCR is about 3000bp, identical to the theoretical length of 3416bp estimated by the designed primer location (promoter to terminator), which could demonstrate that this target plasmid had successfully transformed into E.coli</p><div class="image"><img alt="Colony PCR results of AOX1-&alpha; factor-FMO-AOX1 Terminator, AOX1-&alpha; factor-crtE-AOX1 Terminator, AOX1-&alpha; factor-crtB-AOX1 Terminator and AOX1-&alpha; factor-crtI-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/5/55/T--HUST-China--img--result--4.png" style="width: 100%"/><p>Figure 4: Colony PCR results of AOX1-&alpha; factor-FMO-AOX1 Terminator, AOX1-&alpha; factor-crtE-AOX1 Terminator, AOX1-&alpha; factor-crtB-AOX1 Terminator and AOX1-&alpha; factor-crtI-AOX1 Terminator transformed E.coli</p></div><p>The bands of AOX1-α factor-FMO-AOX1 Terminator (3000+bp), AOX1-α factor-crtE-AOX1 Terminator (almost 3000bp), AOX1-α factor-crtB-AOX1 Terminator (less than 3000bp) and AOX1-α factor-crtI-AOX1 Terminator (3000+bp) from colony PCR are identical to the theoretical lengths of 3214bp, 2746bp, 2767bp and 3316 bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid had successfully transformed into E.coli</p><div class="image"><img alt="Colony PCR results of AOX1-&alpha; factor-CUS-AOX1 Terminator, AOX1-&alpha; factor-ACC-AOX1 Terminator, AOX1-&alpha; factor-4CL-AOX1 Terminator and AOX1-&alpha; factor-LOX2-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/4/46/T--HUST-China--img--result--5.png" style="width: 100%"/><p>Figure 5: Colony PCR results of AOX1-&alpha; factor-CUS-AOX1 Terminator, AOX1-&alpha; factor-ACC-AOX1 Terminator, AOX1-&alpha; factor-4CL-AOX1 Terminator and AOX1-&alpha; factor-LOX2-AOX1 Terminator transformed E.coli</p></div><p>The bands of AOX1-α factor-CUS-AOX1 Terminator (3000bp) , AOX1-α factor-ACC-AOX1 Terminator (3000+bp), AOX1-α factor-4CL-AOX1 Terminator (3000+bp) and AOX1-α factor-LOX2-AOX1 Terminator (almost 5000bp) from colony PCR are identical to the theoretical lengths of 3046bp, 3619bp, 3523bp and 4528bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid had successfully transformed into E.coli</p><div class="image"><img alt="Colony PCR results of AOX1-&alpha; factor-curA-AOX1 Terminator, AOX1-&alpha; factor-pepACS-AOX1 Terminator and AOX1-&alpha; factor-DsbC-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/2/2a/T--HUST-China--img--result--6.png" style="width: 100%"/><p>Figure 6: Colony PCR results of AOX1-&alpha; factor-curA-AOX1 Terminator, AOX1-&alpha; factor-pepACS-AOX1 Terminator and AOX1-&alpha; factor-DsbC-AOX1 Terminator transformed E.coli</p></div><p>The bands of AOX1-α factor-curA-AOX1 Terminator (almost 3000bp), AOX1-α factor-pepACS-AOX1 Terminator (almost 2000bp) and AOX1-α factor-DsbC-AOX1 Terminator (almost 3000bp) from colony PCR are identical to the theoretical lengths of 2875bp, 1987bp and 2722bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid had successfully transformed into E.coli</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h2>The construction of plasmid with Panb1 or Pynr071c as promoter</h2><p>AOX1 promoter is the strongest eukaryotic promoter currently known in yeast expression system. So we choose AOX1 as the primary promoter when we synthesized all these plasmid for the sake of more convenient expression. But noticing that methanol is hazardous, flammable, combustible and therefore, inappropriate to have direct contact with the hair, we need to substrate AOX1 for constitutive promoter Panb1 and xylose induced promoter Pynr071C to realize the projected regulation function as designed. Double-enzyme cleavage and rejointing is used to achieve this. We amplify the target gene located in the primarily synthesized plasmid without AOX1 promoter, and digest the acquired fragments and two kinds of plasmid, containing promoter only, with EcoR I and BamH I, then transform them into E.coli after linking the product together. Through this, we successfully substrate the primary AOX1 for Panb1 and Pynr071C.</p><div class="image"><img alt="Plasmid construction and colony PCR results of reconstructed plasmid with Panb1 and Pynr071C promoter" src="https://static.igem.org/mediawiki/2021/b/b7/T--HUST-China--img--result--7.png" style="width: 100%"/><p>Figure 7: Plasmid construction and colony PCR results of reconstructed plasmid with Panb1 and Pynr071C promoter</p></div><p>All the bands are identical to the theoretical lengths, which could demonstrate that these plasmid are correctly constructed and successfully transformed into E.coli, confirmed by sequencing.</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result2" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result2Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result2" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result2Label">result2</h5></div><div class="modal-body"><p>AOX1 promoter is the strongest eukaryotic promoter currently known in yeast expression system. So we choose AOX1 as the primary promoter when we synthesized all these plasmid for the sake of more convenient expression. But noticing that methanol is hazardous, flammable, combustible and therefore, inappropriate to have direct contact with the hair, we need to substrate AOX1 for constitutive promoter Panb1 and xylose induced promoter Pynr071C to realize the projected regulation function as designed. Another series of plasmid with target gene and new promoter are constructed.</p><div class="image"><img alt="Plasmid construction and colony PCR results of Panb1-&alpha; factor-FMO-AOX1 Terminator and Pynr071c-&alpha; factor-ROX1-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/d/d8/T--HUST-China--img--result--8.png" style="width: 100%"/><p>Figure 8: Plasmid construction and colony PCR results of Panb1-&alpha; factor-FMO-AOX1 Terminator and Pynr071c-&alpha; factor-ROX1-AOX1 Terminator transformed E.coli</p></div><p>The bands of Panb1-α factor-FMO-AOX1 Terminator (less than 3000bp) and Pynr071c-α factor-ROX1-AOX1 Terminator (3000+bp) from colony PCR are identical to the theoretical lengths of 2676bp and 3321bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p><div class="image"><img alt="Plasmid construction and colony PCR results of Panb1-&alpha; factor-4CL-AOX1 Terminator, Panb1-&alpha; factor-crtI-AOX1 Terminator, Panb1-&alpha; factor-crtB-AOX1 Terminator and Panb1-&alpha; factor-crtE-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/9/92/T--HUST-China--img--result--9.png" style="width: 100%"/><p>Figure 9: Plasmid construction and colony PCR results of Panb1-&alpha; factor-4CL-AOX1 Terminator, Panb1-&alpha; factor-crtI-AOX1 Terminator, Panb1-&alpha; factor-crtB-AOX1 Terminator and Panb1-&alpha; factor-crtE-AOX1 Terminator transformed E.coli</p></div><p>The bands of Panb1-α factor-4CL-AOX1 Terminator (3000+bp), Panb1-α factor-crtI-AOX1 Terminator (3000bp), Panb1-α factor-crtB-AOX1 Terminator (2000+bp) and Panb1-α factor-crtE-AOX1 Terminator (2000+bp) from colony PCR are identical to the theoretical lengths of 3185bp, 3046bp, 2198bp and 2177bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p><div class="image"><img alt="Plasmid construction and colony PCR results of Panb1-&alpha; factor-CUS-AOX1 Terminator, Panb1-&alpha; factor-ACC-AOX1 Terminator and Panb1-&alpha; factor-PepACS-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/6/6f/T--HUST-China--img--result--10.png" style="width: 100%"/><p>Figure 10: Plasmid construction and colony PCR results of Panb1-&alpha; factor-CUS-AOX1 Terminator, Panb1-&alpha; factor-ACC-AOX1 Terminator and Panb1-&alpha; factor-PepACS-AOX1 Terminator transformed E.coli</p></div><p>The bands of Panb1-α factor-CUS-AOX1 Terminator (2500bp), Panb1-α factor-ACC-AOX1 Terminator (3000bp) and Panb1-α factor-PepACS-AOX1 Terminator(less than 1500bp) from colony PCR are identical to the theoretical lengths of 2508bp, 3081bp and 1312bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h2>The construction of plasmid without signal peptide</h2><p>During the experiment, we find that not all the protein we want could be secreted extracellularly. Target protein is only detected in the yeast but not in the culture. while no signal appears in the supernatant without them, which indicates that not all of our signal peptides works. We reckon that as the α-factor on the plasmid we used is only a common basic signal peptide, which has some degree of universality but couldn't fit all the protein due to unseen matters like space structure of the protein, our target protein couldn't be induced to secret into the extracellular space by α-factor, or the low expression level coupled with degradation from protease existing in the extracellular environment could do the same. So, to get the rest of our enzymes which don't want to be outside of the cell with α-factor, and whether or not we could raise the expression level, we design new primers for PCR, which eliminate the signal peptide part off the whole target genes, and reconstruct the plasmid without the α-factor to verify if they prefer to be expressed inside of the cell.</p><div class="image"><img alt="Plasmid construction and colony PCR results of plasmid without signal peptides transformed E.coli" src="https://static.igem.org/mediawiki/2021/d/de/T--HUST-China--img--result--11.png" style="width: 100%"/><p>Figure 11: Plasmid construction and colony PCR results of plasmid without signal peptides transformed E.coli</p></div><p>All the bands are identical to the theoretical lengths, which could demonstrate that these plasmid are correctly constructed and successfully transformed into E.coli, confirmed by sequencing.</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result3" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result3Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result3" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result3Label">result3</h5></div><div class="modal-body"><p>Not quite to what we expect, after repeated transfection to the yeast, only a few products are expressed inside of eukaryotic system. Because of the large molecular weight and various types of some of our protein, we suspect that the common signal peptide we use, α-factor, is not enough to bring our protein out of the cell. While there is some of the genes without detectable products and we are hoping to get higher expression level, new primers for PCR are designed to ignore α-factor from our target gene in PCR. Then, likewise, we reconstruct this series of plasmid without α-factor through similar double-enzyme digestion and reconnection which insert our target genes right behind Panb1 promoter.</p><div class="image"><img alt="Plasmid construction and colony PCR results of Panb1-crtB-AOX1 Terminator, Panb1-crtE-AOX1 Terminator and Panb1-FMO-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/5/58/T--HUST-China--img--result--12.png" style="width: 100%"/><p>Figure 12: Plasmid construction and colony PCR results of Panb1-crtB-AOX1 Terminator, Panb1-crtE-AOX1 Terminator and Panb1-FMO-AOX1 Terminator transformed E.coli</p></div><p>The bands of Panb1-crtB-AOX1 Terminator (less than 2000bp), Panb1-crtE-AOX1 Terminator (less than 2000bp) and Panb1-FMO-AOX1 Terminator (2500bp) from colony PCR are identical to the theoretical lengths of 1859bp, 1838bp and 2437bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p><div class="image"><img alt="Plasmid construction and colony PCR results of Panb1-CUS-AOX1 Terminator, Panb1-ACC-AOX1 Terminator, Panb1-4CL-AOX1 Terminator and Panb1-crtI-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/8/8a/T--HUST-China--img--result--13.png" style="width: 100%"/><p>Figure 13: Plasmid construction and colony PCR results of Panb1-CUS-AOX1 Terminator, Panb1-ACC-AOX1 Terminator, Panb1-4CL-AOX1 Terminator and Panb1-crtI-AOX1 Terminator transformed E.coli</p></div><p>The bands of Panb1-CUS-AOX1 Terminator (2000+bp), Panb1-ACC-AOX1 Terminator (3000bp), Panb1-4CL-AOX1 Terminator (2500+bp) and Panb1-crtI-AOX1 Terminator (2500bp) from colony PCR are identical to the theoretical lengths of 2158bp, 2832bp, 2688bp and 2437bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h2>Exploring the construction of plasmid for pigment synthesis</h2><p>After all the expression product is detected, to explore the molecular mechanism laid behind and to save more experience for future teams, we tried to construct the complete whole pathways. Using seamless cloning, tentative experiments are performed by new primer for homologous recombination.</p><p>Unfortunately, no plasmid with multiple target genes are constructed. Good news is double-target-gene plasmid obtained. It seems that the identical promoters and terminators lead to the much shorter product of seamless cloning than theoretical, which means the enzymes used to join the designed homologous arms by primers falsely take some of the shared promoters and terminators as substrates as well. Although FMO dimers were successfully obtained, other plasmids were always shorter than expected .Then we give double-enzyme digestion and reconnection a try but limited to the restriction sites existed in our target genes and more fragments less odds and, well, time, no complete pathway is constructed. What we do have now is the precious experimental experience for all of our team members, and useful information for future teams.</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result4" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result4Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result4" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result4Label">result4</h5></div><div class="modal-body"><div class="image"><img alt="Plasmid construction and colony PCR result of Panb1-&alpha; factor -crtE-AOX1 Terminator-Panb1-&alpha; factor -crtI-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/4/40/T--HUST-China--img--result--14.png" style="width: 100%"/><p>Figure 14: Plasmid construction and colony PCR result of Panb1-&alpha; factor -crtE-AOX1 Terminator-Panb1-&alpha; factor -crtI-AOX1 Terminator transformed E.coli</p></div><p>The bands of Panb1-α factor-crtE-AOX1 Terminator-Panb1-α factor -crtI-AOX1 (less than 5000bp) and is identical to the theoretical lengths of 4574bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p><div class="image"><img alt="Plasmid construction and colony PCR result of Pynr071c-&alpha; factor-FMO dimer-AOX1 Terminator transformed E.coli" src="https://static.igem.org/mediawiki/2021/0/0c/T--HUST-China--img--result--15.png" style="width: 100%"/><p>Figure 15: Plasmid construction and colony PCR result of Pynr071c-&alpha; factor-FMO dimer-AOX1 Terminator transformed E.coli</p></div><p>The bands of FMO dimer (less than 5000bp) and is identical to the theoretical lengths of 4740bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed. After serious of trails, it occurs that when multiple repeated fragments existed, seamless cloning may cause reduction of intended genes and of construction failure. We won't be able to finish the construction of complete pathway when going back for double-enzyme digestion and reconnection, which is much more difficult and complex.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Electroporation and expression of yeast</h1><p>To verify the expression of our genes and to acquire corresponding enzymes, we choose Pichia Pastoris GS115 as chassis and using electroporation to blend our plasmids into them. Large amount of target gene containing plasmid is extracted, then we digest them with Bgl II or Sal I to get linear plasmid, which could be integrated into yeast genome to avoid getting lost while being frozen. Concentration is also applied to achieve higher copy number and higher expression level.</p><p>After electroporation of plasmid with AOX1 as promoter and those without signal peptide, we acquire identical bands through Nickel-affinity chromatography column and SDS-PAGE.</p><p>For some of our enzymes don't have standard protocol to estimate their activity at present, we add substrates into culturing medium accordingly to find out whether there exists active target enzymes and do get our indigo and lycopene synthesized.</p><div class="image"><img alt="Medium for expression with substrates From left to right: GS115 medium with indole and FPP as control; Panb1-FMO-AOX1 Terminator medium with indole; mixture of Panb1-crtE-AOX1 Terminator, Panb1-crtB-AOX1 Terminator, Panb1-crtI-AOX1 Terminator medium with FPP" src="https://static.igem.org/mediawiki/2021/7/79/T--HUST-China--img--result--16.png" style="width: 100%"/><p>Figure 16: Medium for expression with substrates From left to right: GS115 medium with indole and FPP as control; Panb1-FMO-AOX1 Terminator medium with indole; mixture of Panb1-crtE-AOX1 Terminator, Panb1-crtB-AOX1 Terminator, Panb1-crtI-AOX1 Terminator medium with FPP</p></div><p>From left to right: GS115 medium with indole and FPP as control; Panb1-FMO-AOX1 Terminator medium with indole; mixture of Panb1-crtE-AOX1 Terminator, Panb1-crtB-AOX1 Terminator, Panb1-crtI-AOX1 Terminator medium with FPP</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result5" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result5Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result5" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result5Label">result5</h5></div><div class="modal-body"><p>Using E.coli for amplification, we extract and digest them with Bgl II or Sal I to get linear plasmid, which could be integrated into yeast genome to avoid getting lost while being frozen. Then, concentration of linear plasmid is also applied to achieve higher copy number and higher expression level. Several rounds of electroporation later, we successfully get all the plasmid with AOX1 as promoter into yeast.</p><div class="image"><img alt="Colony PCR result of yeast after electroporation" src="https://static.igem.org/mediawiki/2021/f/f3/T--HUST-China--img--result--17.png" style="width: 100%"/><p>Figure 17: Colony PCR result of yeast after electroporation</p></div><p>The bright bands are identical to the theoretical lengths, which could demonstrate that this target plasmid had successfully transformed into yeast.</p><p>After confirmation from colony PCR and sequencing, we using the successfully integrated yeast for expression. At first, we try to detect our target protein in the supernatant since there is signal peptide. At the same time, because the molecular weight of PepACS is only 5.08 kDa, it is almost impossible to separate from bromophenol blue, and its migration is also greatly affected, so the position of the band is not accurate. In order to judge whether it was expressed or not, we first filtered the miscellaneous proteins through 10kDa ultrafiltration tube, then concentrated it with 3kDa ultrafiltration tube, and then detected it by SDS-PAGE. Judge whether it is expressed successfully by whether there is coloring near the bottom line.</p><div class="image"><img alt="SDS-PAGE result of Laccase GS115 4CL LOX2 ACC pepACS DsbC+pepACS detecetion in the supernatant" src="https://static.igem.org/mediawiki/2021/9/9a/T--HUST-China--img--result--18.png" style="width: 100%"/><p>Figure 18: SDS-PAGE result of Laccase GS115 4CL LOX2 ACC pepACS DsbC+pepACS detecetion in the supernatant</p></div><p>Due to glycosylation modification of yeast expression, the molecular weight exhibited on SDS-PAGE will be larger than theoretical. Primary detection shows that we have laccase, 4CL and ACC bands of about 75kDa, LOX2 band of 100+kDa and DsbC+pepACS of about 40kDa, all of which is a bit larger(Laccase: 57.01 kDa; 4CL: 61.88 kDa; ACC: 63.40 kDa; LOX2: 102.88 kDa; DsbC+pepACS: 31.72 kDa) but still within explainable and acceptable range, which could be evidence of successful expression. There are irregular staining bands on the bromophenol blue line in the PepACS swimming lane. Although the molecular weight is not accurate, the successful expression can be judged according to the detection method we designed.</p><p>After confirmation of successful secret of some of the protein, we test the enzymatic activities of laccase and LOX2, which have standard activity testing protocol. Because of the thin band of laccase from SDS-PAGE, we can't be entirely sure whether it's inactive or active but too low the concentration to be detected. So, purification through Nickel-affinity chromatography column is used to raise its concentration for further test of enzymatic activity.</p><div class="image"><img alt="SDS-PAGE result of laccase after purification through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/c/c7/T--HUST-China--img--result--19.png" style="width: 100%"/><p>Figure 19: SDS-PAGE result of laccase after purification through Nickel-affinity chromatography column</p></div><p>Clear and thick band of about 75kDa is consistent to the result in the supernatant. Higher the eluent concentration, thicker the band. This indicates that the band of laccase in Fig18 is our target protein instead of other impurity with a high expression level.</p><p>During the test, we find a vague band of ROX1 which couldn't be verified as successfully expressed according to its SDS-PAGE result in the supernatant. As the key component of our xylose responding system, the successful expression of ROX1 is most vital to our project, so we apply purification through Nickel-affinity chromatography column to ROX1 to further identify its expression. Lucky us, target band is acquired and confirmed. It feels so good to announce that our ROX1 can be expressed like the others.</p><div class="image"><img alt="SDS-PAGE result of ROX1 after purification through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/2/21/T--HUST-China--img--result--20.png" style="width: 100%"/><p>Figure 20: SDS-PAGE result of ROX1 after purification through Nickel-affinity chromatography column</p></div><p>Different from impure or permeate bands, the target protein located around 55-55kDa, bigger than the theoretical 47.09kDa but still within explainable and acceptable range of glycosylation modification. ROX1 could be confirmed as successfully expressed. Although no gradient elution is applied, following elution result could verify this is ROX1, with some impurity bands occurring in the first ROX1 elution.</p><p>At the same time, after several SDS-PAGE confirmation, some of our proteins still don't want to show themselves. No band detected. We infer that this is due to A. the low level of extracellular expression and degradation by protease existing in the extracellular environment and B. the common signal peptide we use-α-factor-isn't specialized according to the unique space structure of our proteins so the mismatch causes failure of extracellular expression. To solve this, we reconstruct plasmids without the signal peptide and try to do intracellular expression. This is aim at all the undetectable or low-expressed genes.</p><div class="image"><img alt="Colony PCR result of yeast after electroporation of reconstructed plasmid without the signal peptide" src="https://static.igem.org/mediawiki/2021/d/db/T--HUST-China--img--result--21.png" style="width: 100%"/><p>Figure 21: Colony PCR result of yeast after electroporation of reconstructed plasmid without the signal peptide</p></div><p>The bright bands are identical to the theoretical lengths, which could demonstrate that this target plasmid had successfully transformed into yeast. Target genes are confirmed exist in the yeast of multiple bands, which could be the result of polluted electroporation cup.</p><p>After verification of successful transfection, we can't test the protein directly due to intracellular expression. So, we extract the total protein in yeast and go for a purification through Nickel-affinity chromatography column, then apply SDS-PAGE to separate target protein from the large amount and various type of total protein to confirm whether our target protein could be expressed and value its expression level quantitatively.</p><div class="image"><img alt="SDS-PAGE result of FMO after purification of yeast total protein extraction product through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/e/ed/T--HUST-China--img--result--22.png" style="width: 100%"/><p>Figure 22: SDS-PAGE result of FMO after purification of yeast total protein extraction product through Nickel-affinity chromatography column</p></div><p>Different from impure or permeate bands, the target protein located around 60kDa, bigger than the theoretical 53.96kDa but still within explainable and acceptable range of glycosylation modification. FMO could be confirmed as successfully expressed. The concentration of yeast total protein is so high that huge amount of impure protein is included during elution. But due to difference from impure or permeate bands, its dark color and consistency among several times of elution, this band could be verified as our target FMO,</p><div class="image"><img alt="SDS-PAGE result of crtE after purification of yeast total protein extraction product through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/1/16/T--HUST-China--img--result--23.png" style="width: 100%"/><p>Figure 23: SDS-PAGE result of crtE after purification of yeast total protein extraction product through Nickel-affinity chromatography column</p></div><p>Different from impure or permeate bands, the target protein located around 50kDa, bigger than the theoretical 33.42kDa but still within explainable and acceptable range of glycosylation modification. crtE could be confirmed as successfully expressed.</p><div class="image"><img alt="SDS-PAGE result of crtB after purification of yeast total protein extraction product through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/f/fc/T--HUST-China--img--result--24.png" style="width: 100%"/><p>Figure 24: SDS-PAGE result of crtB after purification of yeast total protein extraction product through Nickel-affinity chromatography column</p></div><p>Different from impure or permeate bands, the target protein located around 50kDa, bigger than the theoretical 35.30kDa but still within explainable and acceptable range of glycosylation modification. crtB could be confirmed as successfully expressed.</p><div class="image"><img alt="SDS-PAGE result of crtI after purification of yeast total protein extraction product through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/6/6f/T--HUST-China--img--result--25.png" style="width: 100%"/><p>Figure 25: SDS-PAGE result of crtI after purification of yeast total protein extraction product through Nickel-affinity chromatography column</p></div><p>Different from impure or permeate bands, the target protein located around 60kDa, bigger than the theoretical 55.86kDa but still within explainable and acceptable range of glycosylation modification. crtI could be confirmed as successfully expressed. After confirmation of successful expression, we add indole as substrate into culture medium of FMO and FPP into the mixture of crtE, crtB and crtI to test whether the expressed enzyme is active in terms of color change of the culture medium. As for the result, we succeed in expression of enzymes and synthesis of indigo and lycopene.</p><div class="image"><img alt="Medium for expression with substrates From left to right: GS115 medium with indole and FPP as control; Panb1-FMO-AOX1 Terminator medium with indole; mixture of Panb1-crtE-AOX1 Terminator, Panb1-crtB-AOX1 Terminator, Panb1-crtI-AOX1 Terminator medium with FPP" src="https://static.igem.org/mediawiki/2021/0/03/T--HUST-China--img--result--26.png" style="width: 100%"/><p>Figure 26: Medium for expression with substrates From left to right: GS115 medium with indole and FPP as control; Panb1-FMO-AOX1 Terminator medium with indole; mixture of Panb1-crtE-AOX1 Terminator, Panb1-crtB-AOX1 Terminator, Panb1-crtI-AOX1 Terminator medium with FPP</p></div><p>From left to right:</p><p>GS115 medium with indole and FPP as control; Panb1-FMO-AOX1 Terminator medium with indole; mixture of Panb1-crtE-AOX1 Terminator,Panb1-crtB-AOX1 Terminator,Panb1-crtI-AOX1 Terminator medium with FPP</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><p>Also, In order to verify whether the catalytic effect of FMO dimer is better than FMO. We electrotransformed the FMO dimer into yeast and tried to detect its expression and compare it with FMO.</p><p>The color of the medium and the cell were significantly darker than FMO under the same culture conditions. This shows that the catalytic activity of FMO dimer is stronger than that of FMO.</p><div class="image"><img alt="FMO dimer medium with indole after centrifugation.It is obvious that the culture medium and bacteria are dark blue." src="https://static.igem.org/mediawiki/2021/3/3a/T--HUST-China--img--result--27.png" style="width: 100%"/><p>Figure 27: FMO dimer medium with indole after centrifugation.It is obvious that the culture medium and bacteria are dark blue.</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result6" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result6Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result6" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result6Label">result6</h5></div><div class="modal-body"><p>After electrotransformed the FMO dimer into yeast, we still used Colony PCR to confirm the target gene is successfully transformed into the yeast cells.</p><div class="image"><img alt="Colony PCR result of yeast after electroporation of FMO dimer." src="https://static.igem.org/mediawiki/2021/e/ef/T--HUST-China--img--result--28.png" style="width: 100%"/><p>Figure 28: Colony PCR result of yeast after electroporation of FMO dimer.</p></div><p>The bands of FMO dimer (less than 5000bp) and is identical to the theoretical lengths of 4600bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that this target plasmid had successfully transformed into yeast.</p><p>At first, we tried to detect the protein in the supernatant, but no results were obtained. Because we have already experienced similar problems. We extracted the total protein directly and go for a purification to tested whether it was in the cell. Actually we detected the protein this time but it was smaller than expected.</p><div class="image"><img alt="SDS-PAGE result of FMO dimer after purification of yeast total protein extraction product through Nickel-affinity chromatography column" src="https://static.igem.org/mediawiki/2021/e/e6/T--HUST-China--img--result--29.png" style="width: 100%"/><p>Figure 29: SDS-PAGE result of FMO dimer after purification of yeast total protein extraction product through Nickel-affinity chromatography column</p></div><p>Different from impure or permeate bands, the target protein located around 60kDa, smaller than the theoretical 107.52kDa, but similar to the theoretical molecular weight of FMO (53.96kDa).</p><p>This indicated that the FMO dimer was broken in the process of expression or extraction. In order to confirm whether the catalytic effect is better, we also added indole to its medium to observe its effect on indigo synthesis.</p><div class="image"><img alt="FMOdimer medium with indole after centrifugation" src="https://static.igem.org/mediawiki/2021/e/ef/T--HUST-China--img--result--28.png" style="width: 100%"/><p>Figure 30: FMOdimer medium with indole after centrifugation</p></div><p>It is obvious that the culture medium and bacteria are dark blue.</p><p>The color of the medium and the cell were significantly darker than FMO under the same culture conditions. This shows that the catalytic activity of FMO dimer is stronger than that of FMO. So we can confirm that the expression is FMO dimer, and its effect is better. As for the problem of suspected fracture, we suspect that it is due to the short connection. It will withstand a large torsional strain when folding, resulting in its fragility, and fracture occurs in the process of extraction or sample preparation.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Determination of enzyme avtivity</h1><p>After the target strips appeared on SDS-PAGE, for the enzymes with standard enzyme activity assay such as Laccase and LOX2, we measured their enzyme activity according to the standard measurement methods to detect whether our enzymes are catalytically active. Laccase and LOX2 show high activity after adjusting the relevant conditions such as pH, temperature and ion concentration</p><div class="image"><img alt="Enzyme activity determination of Laccase" src="https://static.igem.org/mediawiki/2021/f/f2/T--HUST-China--img--result--31.png" style="width: 100%"/><p>Figure 31: Enzyme activity determination of Laccase</p></div><div class="image"><img alt="Enzyme activity determination of LOX2" src="https://static.igem.org/mediawiki/2021/0/02/T--HUST-China--img--result--32.png" style="width: 100%"/><p>Figure 32: Enzyme activity determination of LOX2</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result7" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result7Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result7" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result7Label">result7</h5></div><div class="modal-body"><p>After target strips been detected, to measure the activity of Laccase, we mixed 1ml of 1mmol / L ABTS solution with 1ml of centrifuged supernatant and adjusted the final concentration of Cu2+ to 10mmol/L, and measured its absorbance at 420nm. On this basis, we carried out determination experiments to further explore the optimal conditions for inducing the highest activity of Laccase.</p><p>Since the active center of Laccase is copper ion, we first explored the copper ion concentration to be added into the culture media. Adjusted the concentration of supplementary CuSO4 solution to 9mmol / L and 10mmol / L respectively, and measured the enzyme activity of Laccase under the same conditions</p><div class="image"><img alt="Effects of different concentrations of CuSO4 on enzyme activity" src="https://static.igem.org/mediawiki/2021/7/79/T--HUST-China--img--result--33.png" style="width: 100%"/><p>Figure 33: Effects of different concentrations of CuSO4 on enzyme activity</p></div><p>We found that the enzyme activity reaches its zenith when the concentration of copper ion is 10mmol/L. It is probably because the concentration of Cu2+ can't meet the need of Laccase active center when it is too low. When the concentration is too high, it will affect the binding of Laccase and substrate due to the existence of too much copper ions.</p><p>We also explored the effects of pH and temperature on activity of Laccase at the same time.</p><p>The optimum pH of Laccase was found to be acidic, therefore we prepared buffers with pH = 3, pH = 4.8 and pH = 6.6 respectively, with 100 mM citric acid and sodium citrate and added 1 ml buffer to control the pH of the reaction system while maintaining the final concentration of CuSO4 at 10 mmol / L</p><div class="image"><img alt="Effects of different pH on enzyme activity" src="https://static.igem.org/mediawiki/2021/a/a0/T--HUST-China--img--result--34.png" style="width: 100%"/><p>Figure 34: Effects of different pH on enzyme activity</p></div><p>After comparison, we found that among the three groups of data measured, Laccase activity is highest at pH = 3.0, decreasing to nadir at pH = 4.8, and completely inactivated at pH = 6.6. As for the reason why the subsequent measured value was lower than that at the beginning at pH 6.6, we speculated that it was caused by undermixing of solution when started reading.</p><p>For the effect of temperature, we kept ABTS solution and supernatant at 10℃, 20℃ and 37℃ respectively in advance, and kept the final concentration of CuSO4 at 10mmol / L.</p><div class="image"><img alt="" src="https://static.igem.org/mediawiki/2021/6/6a/T--HUST-China--img--result--35.png" style="width: 100%"/><p>Figure 35:</p></div><p>Through the analysis and comparison of the three groups of data, it is found that under testing conditions, the activity of Laccase is the highest when the temperature is 20℃, and the Laccase activity is inhibited by both low and high temperatures. As for the decrease of absorbance at the final phase of measuring enzyme activity at 10℃, it is suspected that the low temperature has a certain impact on ABTS cationic free radical, resulting in the decrease in absorbance</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Verification of promoters</h1><p>As we had successfully used Panb1 as promoter to express our protein, which can confirm that the Panb1 promoter can work smoothly. So to verify whether our Pynr071c promoter could initiate the expression of downstream genes, we linked GFP it, then transfect it into yeast and test the exist of fluorescence.</p><div class="image"><img alt="Fluorescence result of Pynr071c-GFP-AOX1 Terminator" src="https://static.igem.org/mediawiki/2021/d/d5/T--HUST-China--img--result--36.png" style="width: 100%"/><p>Figure 36: Fluorescence result of Pynr071c-GFP-AOX1 Terminator</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result8" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result8Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result8" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result8Label">result8</h5></div><div class="modal-body"><p>To verify whether our Pynr071c promoter could initiate the expression of downstream genes, we linked GFP it, then transfect it into yeast and test the exist of fluorescence.</p><div class="image"><img alt="Plasmid construction and colony PCR results of Pynr071c-GFP-AOX1 Terminator transformed E.coli" style="width: 100%"/><p>Figure 37: Plasmid construction and colony PCR results of Pynr071c-GFP-AOX1 Terminator transformed E.coli</p></div><p>The bands of Pynr071c-GFP-AOX1 Terminator from colony PCR are identical to the theoretical lengths of 2266bp estimated by the designed primer locations (promoter to terminator), which could demonstrate that these target plasmid are successfully constructed.</p><p>We successfully construct the plasmid with the GFP and sequencing of which is correct. E.coli is used for amplification as well. Extract plasmid from E.coli, digest with Bgl II to get linear plasmid and concentrate for electroporation of yeast.</p><div class="image"><img alt="Colony PCR result of yeast after electroporation. The bright bands are identical to the theoretical lengths, which could demonstrate that this target plasmid had successfully transformed into yeast." src="https://static.igem.org/mediawiki/2021/3/34/T--HUST-China--img--result--38.png" style="width: 100%"/><p>Figure 38: Colony PCR result of yeast after electroporation. The bright bands are identical to the theoretical lengths, which could demonstrate that this target plasmid had successfully transformed into yeast.</p></div><p>We induce the plasmid with confirmation of colony PCR to express. Adding xylose everyday for xylose induced promoter Pynr071C to induce synthesis of GFP. Using fluorescence microscopy to detect the exist of green fluorescence, which indicates whether our promoters function regularly.</p><div class="image"><img alt="Fluorescence result of Pynr071c-GFP-AOX1 Terminator" src="https://static.igem.org/mediawiki/2021/4/42/T--HUST-China--img--result--39.png" style="width: 100%"/><p>Figure 39: Fluorescence result of Pynr071c-GFP-AOX1 Terminator</p></div></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>The Experiment Standard</h1><p>We measured the standard curves of three pigments before using them for hair dyeing experiment. We also found that the amount of melanin contained in hair can have a significant effect on hair dyeing outcomes. Therefore, we define different colors of hair based on bleaching.</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result1" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result1Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result1" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result1Label">result1</h5></div><div class="modal-body"><div class="image"><img alt="Curcumin concentration standard curve" src="https://static.igem.org/mediawiki/2021/8/89/T--HUST-China--img--result--result-curcumin_concentration_sc.png" style="width: 100%"/><p>Figure 40: Curcumin concentration standard curve</p></div><div class="image"><img alt="Indigo concentration standard curve" src="https://static.igem.org/mediawiki/2021/4/42/T--HUST-China--img--result--result-Indigo_concentration_sc.png" style="width: 100%"/><p>Figure 41: Indigo concentration standard curve</p></div><div class="image"><img alt="sLycopene concentration standard curve" src="https://static.igem.org/mediawiki/2021/5/55/T--HUST-China--img--result--result-Lycopene_concentration_sc.png" style="width: 100%"/><p>Figure 42: sLycopene concentration standard curve</p></div><div class="image"><img alt="Series of bleached hair" src="https://static.igem.org/mediawiki/2021/e/e9/T--HUST-China--img--result--result-the_sample_from_shop.png" style="width: 100%"/><p>Figure 43: Series of bleached hair</p></div><p>For the convenience of dyeing, we buy a series of bleached hair. The lightest color is the 9°hair, and the darkest is the 4 ° hair.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>The Best Condition of hair dye</h1><p>We have gained the best dye conditions of three kinds of hair dye(indigo, curcumin and lycopene) at a certain concentration. Under optimal conditions, we dyed 4-9 degrees of hair to get a series of dyeing discs. And we found that as for the three colors selected for the experiment, bleach the hair to 8 degrees could achieve a bright coloring effect.</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result2" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result2Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result2" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result2Label">result2</h5></div><div class="modal-body"><p>Chart of the best condition of hair dye</p><table><thead><tr><th>Dye/Condition</th><th>temperature</th><th>Dyeing aid ingredients</th><th>concentration(g/L)</th><th>comment</th></tr></thead><tbody><tr><td>indigo 2min</td><td>Room temperature</td><td>none</td><td>2</td><td>The color deepens significantly while dyeing for multiple times</td></tr><tr><td>curcumin 30min</td><td>50℃</td><td>Ethyl alcohol</td><td>0.5</td><td></td></tr><tr><td>lycopene 30min</td><td>Room temperature</td><td>alum</td><td>2</td><td></td></tr></tbody></table><p><em>The best dyeing condition</em></p><p>The exhibition of hair dyed on the best condition</p><p>Under the best conditions, we dyed the hair from 4 degree to 9 degree, and got a series of colors. It is found that it only needed to be bleached to 8 degree so that the hair would show a bright color for all three kinds of dye.</p><p>As to lycopene hair, 8 or 9 degree hair was red, 7 degree(or below) hair was brownish, and the longer the hair was dyed, the redder it would become.</p><p>As to indigo hair, 7 to 9 degree hair would become blue. As the dyeing time goes, the color would turn blue from an indigo color; 5 to 6 degree hair would be dyed to celadon, and 4 degree hair was still brown.</p><div class="image"><img alt="The dyeing results of lycopene(room temperature, adding alum, 2g/L). From left to right: 9&#176;(5,10,30min), 8&#176;(5,10,30min), 7&#176;(5,10,30min), 6&#176;(5,10,30min), 5&#176;(10,30min), 4&#176;(5,10,30min)" src="https://static.igem.org/mediawiki/2021/9/92/T--HUST-China--img--result--result-the_best_condition_for_lycopene_to_dye_.png" style="width: 100%"/><p>Figure 44: The dyeing results of lycopene(room temperature, adding alum, 2g/L). From left to right: 9&#176;(5,10,30min), 8&#176;(5,10,30min), 7&#176;(5,10,30min), 6&#176;(5,10,30min), 5&#176;(10,30min), 4&#176;(5,10,30min)</p></div><div class="image"><img alt="The dyeing results of indigo (room temperature, 2g/L). From left to right: 9&#176;(0.5,2,6min), 8&#176;(0.5,2,6min), 7&#176;(0.5,2,6min), 6&#176;(0.5,2,6min), 5&#176;(0.5,2,6min), 4&#176;(0.5,2,6min)" src="https://static.igem.org/mediawiki/2021/8/89/T--HUST-China--img--result--result-the_best_condition_for_indigo_to_dye.png" style="width: 100%"/><p>Figure 45: The dyeing results of indigo (room temperature, 2g/L). From left to right: 9&#176;(0.5,2,6min), 8&#176;(0.5,2,6min), 7&#176;(0.5,2,6min), 6&#176;(0.5,2,6min), 5&#176;(0.5,2,6min), 4&#176;(0.5,2,6min)</p></div><div class="image"><img alt="The dyeing results of indigo(room temperature, ethanol added, 0.5g/L). From left to right: 10min(9-4&#176;), 30min(9-4&#176;)8&#176;" src="https://static.igem.org/mediawiki/2021/e/e4/T--HUST-China--img--result--result-the_best_condition_for_turmeric_to_dye.png" style="width: 100%"/><p>Figure 46: The dyeing results of indigo(room temperature, ethanol added, 0.5g/L). From left to right: 10min(9-4&#176;), 30min(9-4&#176;)8&#176;</p></div><p>Problems occurred during the research:</p><p><strong>Lycopene</strong></p><p>Problem: No literature on coloring fabrics or hair with lycopene</p><p>Solution: We conducted a gradient experiment (0.5, 1, 2, 5 g/L) to explore the effective concentration of lycopene for hair dyeing. Finally, 2g/L of lycopene is selected, at which concentration the dye fluid will not be too viscous, and has a better dyeing effect as the picture below shows.</p><div class="image"><img alt="(From left to right: 0.5(30 min), 1(5, 10, 30 min), 2(5, 10, 30min), 5(5, 10, 30min)g/L of lycopene)" src="https://static.igem.org/mediawiki/2021/3/35/T--HUST-China--img--result--result-the_effort_to_confirm_the_best_concentration_for_lycopene_dying.png" style="width: 100%"/><p>Figure 47: (From left to right: 0.5(30 min), 1(5, 10, 30 min), 2(5, 10, 30min), 5(5, 10, 30min)g/L of lycopene)</p></div><p>Problem: Lycopene dye the hair with a low efficiency, a low color fastness, and a constantly discoloring process when the hair is showered by water.</p><p>Solution: We looked up the data and selected three eco-friendly color aids (alum, potassium tartrate, citric acid). Through direct color comparison and elution experiments, we found that alum can significantly improve the coloration rate and color fastness of lycopene.</p><div class="image"><img alt="(From left to right, 1st-4th groups: alum(30min), potassium tartrate(40min), citric acid(40 min), no color aids(40min); 5th-8th groups: the 1st-4th hair after washing 7 times)" src="https://static.igem.org/mediawiki/2021/6/6d/T--HUST-China--img--result--result-the_effort_for_confirming_the_maintaining_degree_of_lycopene.png" style="width: 100%"/><p>Figure 48: (From left to right, 1st-4th groups: alum(30min), potassium tartrate(40min), citric acid(40 min), no color aids(40min); 5th-8th groups: the 1st-4th hair after washing 7 times)</p></div><div class="image"><img alt="Color fastness test of lycopene" src="https://static.igem.org/mediawiki/2021/c/ca/T--HUST-China--img--result--result-Color_fastness_test_of_lycopene.png" style="width: 100%"/><p>Figure 49: Color fastness test of lycopene</p></div><p><strong>Curcumin</strong></p><p>Problem: The coloration rate of the curcumin aqueduct solution is low</p><p>Solution: We carried out the same dye addition and elution experiments as lycopene, and found that alum, potassium tartarate and citric acid cannot improve the coloring effect of curcumin. The data showed that curcumin is soluble in ethanol, and the optimal coloring temperature is 50 degree, so we adjusted the solvent to 33% ethanol solution. The process of coloring was conducted in the oven at 50 degrees, and the results showed that the coloring effect significantly improved.</p><div class="image"><img alt="Dyeing result of curcumin made in different solvents(30 min, 50&#176;C). Left: water solution, Right: 33% ethanol alcohol solution" src="https://static.igem.org/mediawiki/2021/c/c6/T--HUST-China--img--result--result-Dyeing_result_of_curcumin_made_in_different_solvents.png" style="width: 100%"/><p>Figure 50: Dyeing result of curcumin made in different solvents(30 min, 50&#176;C). Left: water solution, Right: 33% ethanol alcohol solution</p></div><p><strong>Indigo</strong> Problem: At the beginning of the experiment, we found that indigo could not be successfully adsorbed by the hair.</p><p>Solution: We looked up the data and found that in the printing and dyeing industry, it is necessary to restore the low solubility of indigo to water-soluble leucoindigo, which is used to color. We used the textile reducing agent - sodium sulfate to reduce indigo dye, and it came out to be successful.</p><p>Considering the safety of hair dyeing, we tried to use glucose as a reducing agent, by which we also obtained effective dyeing fluids. The method of using is indigo: glucose: caustic soda: 1:6:1, react for 15 min under 50°C. In the final product, we expected that yeast could conduct oxygen-free fermentation to reduce indigo with ethanol.</p><p>Problem: After dyeing your hair indigo, your hair becomes noticeably brittle.</p><p>Solution: Testing the average pull limit of a single hair, we found that the hair pull limit for indigo treatment was indeed significantly lower than that of normal hair. We assumed that this is related to the destruction of hair by alkaline substances. Therefore, we shorten the processing time of indigo dye, and finally designed to dye 2min at a time, repeated the procedure five times to achieve a saturation effect.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Hair stress experiment</h1><p>We used a micro scale experimental instrument to quantitatively evaluate the changes in mechanical properties of hair after hair dyeing. The results show that Indigo and lycopene as dyes have no obvious damage to hair. Curcumin has a repairing effect on hair.</p><div class="image"><img alt="Tensile Stress Strain Curves" src="https://static.igem.org/mediawiki/2021/e/ec/T--HUST-China--img--result--result-Tensile_Stress_Strain_Curves.png" style="width: 100%"/><p>Figure 51: Tensile Stress Strain Curves</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result3" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result3Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result3" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result3Label">result3</h5></div><div class="modal-body"><p>After hair dyeing, the mechanical properties of hair in each group changed to varying degrees. For example, the hair softens after a long treatment of the indigo solution becomes soft, elastic but easy to break. So we borrowed the micro scale experimental instrument independently developed by Professor Liu.</p><p>During the pre-experiment, we found there is no significant difference in ultimate stress between traditional dyeing hair and the untreated one, and we believe it's the added hair repair component causes the slug. Indigo and lycopene as dyes also have no obvious damage to hair. What's more, the yang's modulus of curcumin treatment hair is significantly higher. So we believe it keeps hair tough.</p><div class="image"><img alt="Indigo dyed hair being treated with micro tensile / micro torsional mechanical tester" src="https://static.igem.org/mediawiki/2021/b/b5/T--HUST-China--img--result--result-Indigo_dyed_hair_being_treated_with_micro_tensile_micro_torsional_mechanical_tester.png" style="width: 100%"/><p>Figure 52: Indigo dyed hair being treated with micro tensile / micro torsional mechanical tester</p></div></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Compound hair dye paste</h1><p>After finishing the solution experiment, we try to mix the natural pigment into a dye that can be applied directly to the hair. At present, lycopene dye and curcumin dye with NO.1 cream matrix as carrier are obtained, and natural essence is added to improve the odor of dye paste. Indigo is an oxidizing dye with special properties, so we designed a timely fermenter. In this way, we can use our product right now when indigo is produced and reduced to indigo white.</p><div class="image"><img alt="The expected product" src="https://static.igem.org/mediawiki/2021/5/55/T--HUST-China--img--result--result-the_expected_product.png" style="width: 100%"/><p>Figure 53: The expected product</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result4" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result4Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result4" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result4Label">result4</h5></div><div class="modal-body"><p><strong>Lycopene</strong></p><p>Difficulty: when we use lycopene paste to dye hair, it is not red but orange</p><p>Solution: by analyzing the cream formula, we think what is causing this problem is sodium sulfite. We add sodium sulfite to prevent further oxidation of the pigment, but it may also reduce and fade the pigment. The solution experiment proved our conjecture. A new lycopene dye that doesn't contain sodium sulfite found its way to red hair.</p><p><strong>Lycopene dying cream</strong></p><table><thead><tr><th>Ingredient</th><th>Content</th></tr></thead><tbody><tr><td>Cream matrix</td><td>100g</td></tr><tr><td>Sodium sulfite</td><td>0.2g</td></tr><tr><td>Absolute ethanol</td><td>1mL</td></tr><tr><td>pH 6.8 phosphate buffer</td><td>1mL</td></tr><tr><td>Solid paraffin</td><td>1 drop or not</td></tr><tr><td>Essence</td><td>1 drop</td></tr><tr><td>20% Lycopene</td><td>5g</td></tr><tr><td>Alum</td><td>0.6g</td></tr></tbody></table><p><strong>Curcumin</strong> EDTA is added to the curcumin paste to prevent metal ions from interfering with the effect -- for example, Fe3 + producing a reddish-brown tint.</p><p><strong>Curcumin dying cream</strong></p><table><thead><tr><th>Ingredient</th><th>Content</th></tr></thead><tbody><tr><td>Cream matrix</td><td>100g</td></tr><tr><td>EDTA</td><td>0.2g</td></tr><tr><td>Sodium sulfite</td><td>0.1g</td></tr><tr><td>Absolute ethanol</td><td>1mL</td></tr><tr><td>pH 6.8 phosphate buffer</td><td>1mL</td></tr><tr><td>Isopropyl alcohol</td><td>2mL</td></tr><tr><td>Solid paraffin</td><td>1 drop or not</td></tr><tr><td>Essence</td><td>1 drop</td></tr><tr><td>curcumin</td><td>0.5g</td></tr></tbody></table><div class="image"><img alt="The result of curcumin staining cream. 9 &#176; from left to right. Hair was dyed at room temperature for 30min, 60min and 0min; Dyeing at 50 &#8451; for 10min and 30min" src="https://static.igem.org/mediawiki/2021/a/a0/T--HUST-China--img--result--result-the_different_effort_of_termeric_dying_at_changing_tep.%26time.png" style="width: 100%"/><p>Figure 54: The result of curcumin staining cream. 9 &#176; from left to right. Hair was dyed at room temperature for 30min, 60min and 0min; Dyeing at 50 &#8451; for 10min and 30min</p></div><p>The result of curcumin staining cream. 9 ° from left to right. Hair was dyed at room temperature for 30min, 60min and 0min; Dyeing at 50 ℃ for 10min and 30min</p><p><strong>Indigo:</strong></p><p>Difficult: we can make indigo paste, but the hair does not dye well.</p><p>Solution: Indigo is a water-soluble component, and need to be oxidized to indigo after fixing to the hair. With water-in-oil paste as the matrix, indigo white can not fully enter the interior of the hair, and the oily substances in the matrix and excessive reductant prevent indigo white from oxidation in the hair, resulting in no effective coloring.</p><p>So we decided to design a timely manner in which indigo could be produced and used at the same time. Therefore, we consider that indigo dye can be produced and used in time -- the direct production of indigo by yeast, and the production of indigo solution as a dye in time.</p><p>For this idea, we dye indigo solution directly on the hair and find that it can be painted, but it can not color the hair evenly. So we designed a hair dye comb to make it possible to evenly smear indigo cryptosomes on the hair. The matching device is a timely fermentation tank, which can meet the needs of users with our engineering bacteria as raw materials, timely production and timely use of indigo white. For detailed information, please refer to <a href="https://2021.igem.org/Team:HUST-China/Results/Hardware">Hardware</a> part.</p><div class="image"><img alt="the different effort of indigo" src="https://static.igem.org/mediawiki/2021/5/5b/T--HUST-China--img--result--result-the_different_effort_of_indigo_dying_at_changing_tep.%26time.png" style="width: 100%"/><p>Figure 55: the different effort of indigo</p></div></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Color fastness test</h1><p>Color fastness is an important aspect to measure the effect of dye, so we design a set of elution scheme and test the color fastness of three kinds of natural pigment dye products and the same color traditional dye paste. The results showed that the color fastness of the natural pigment dyes was better than that of the traditional dyes.</p><div class="image"><img alt="The comparison of biological dying with traditional dying" src="https://static.igem.org/mediawiki/2021/e/ee/T--HUST-China--img--result--result-the_comparison_of_biological_dying_with_traditional_dying.png" style="width: 100%"/><p>Figure 56: The comparison of biological dying with traditional dying</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result5" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result5Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result5" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result5Label">result5</h5></div><div class="modal-body"><p>The experiment of color fastness:</p><p>Compound 25g/L shampoo solution. Add 10 ML shampoo solution and a bunch of hair in a 15 ML centrifuge tube for 5 min at 100 r/min. The experiment was repeated seven times. The absorbance of the eluent at the maximum absorption band of the corresponding color was measured and the curve was drawn. Results as shown in figure, the single elution amount of three kinds of natural pigment was less than that of the same color traditional dyes, and the color change after 7 times elution was also less than that of the traditional dyes. Furthermore, curcumin dyeing cream treatment of hair is almost non-decolorization.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Toxicological experiment of broad bean root tip</h1><p>In order to test the safety of our dye paste, we carried out a toxicology experiment of broad bean root tip. The rate of micronucleus in root tip cells on broad bean was measured. The higher the micronucleus rate was, the higher the teratogenic rate was. The results showed that the teratogenicity of our product was significantly lower than that of the two traditional hair dyes on the market, indicating that our product was less toxic.</p><div class="image"><img alt="Micronucleus Frequency" src="https://static.igem.org/mediawiki/2021/0/0a/T--HUST-China--img--result--result-micronucleus_frequency.png" style="width: 100%"/><p>Figure 57: Micronucleus Frequency</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result6" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result6Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result6" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result6Label">result6</h5></div><div class="modal-body"><p>Micronucleus (MCN) is an abnormal structure in eukaryotic cells, which is often produced by the action of radiation or chemical drugs. In the intercellular phase, the Micronuclei were round or oval, free from the main nucleus, the size should be less than 1/3 of the main nucleus. Only the eucaryotic test system can directly predict the genetic damage of mutagens to human beings and other higher organisms. In this respect, the micronucleus test is an ideal method. And some studies show that the plant micronucleus test and the animal test have a coincidence rate of up to 99%. Micronucleus test using broad bean root tip as experimental material can accurately show the effect of various treatments on induced distortion.</p><div class="image"><img alt="Patricles" src="https://static.igem.org/mediawiki/2021/d/d7/T--HUST-China--img--result--result-patricles.png" style="width: 100%"/><p>Figure 58: Patricles</p></div><p>Specific experimental process:</p><ol><li>We bought 2 catties of sun-dried broad bean seeds, put them in a wet porcelain tray after soaking in water overnight. After 2 days, about 95% of the bean seeds grew roots about 1-2 cm long.</li><li>We divided 10 broad beans into 1 group. Each group of root tips were immersed in the following concentrations of dye paste solution for 24 hours.</li><li>We took the broad beans soaked in the dye solution, washed them with distilled water, then put them into distilled water and cultured for 24 hours.</li><li>The root tips of Vicia Faba were cut off and fixed in 15-fold Kano Solution (anhydrous ethanol: Glacial Acetic acid = 3:1) for 24 hours.</li><li>We took out the root tip, washed it with distilled water, put it into 1 m hydrochloric acid solution, and reacted at 60 °C for 20 minutes to soften the root tip. The root tip was cut 1 mm, smashed on the slide, stained with the modified carbolic acid red dye for 15 min, covered with the slide, then flattened and observed under the microscope.</li><li>We first find the meristematic cells, which are closely arranged and square, look at 1000 cells, and record the number of cells with micronucleus. Micronucleus rate = micronucleus cell number/1000 ?? 1000%</li></ol><table><thead><tr><th>Indigo</th><th>Traditional blue</th><th>Lycopene</th><th>Traditional red</th><th>curcumin</th><th>Negative control:Distilled water</th><th>Positive control:1g/L CuSO4</th></tr></thead><tbody><tr><td>9g/L</td><td>9g/L</td><td>9g/L</td><td>9g/L</td><td>9g/L</td><td></td><td></td></tr><tr><td>6g/L</td><td>6g/L</td><td>6g/L</td><td>6g/L</td><td>6g/L</td><td></td><td></td></tr><tr><td>4.5g/L</td><td>4.5g/L</td><td>4.5g/L</td><td>4.5g/L</td><td>4.5g/L</td><td></td><td></td></tr><tr><td>3g/L</td><td>3g/L</td><td>3g/L</td><td>3g/L</td><td>3g/L</td><td></td><td></td></tr><tr><td>1.5g/L</td><td>1.5g/L</td><td>1.5g/L</td><td>1.5g/L</td><td>1.5g/L</td><td></td><td></td></tr></tbody></table><div class="image"><img alt="Broad bean dyed with pigment" src="https://static.igem.org/mediawiki/2021/7/70/T--HUST-China--img--result--result-broad_bean_dyed_with_pigment.png" style="width: 100%"/><p>Figure 59: Broad bean dyed with pigment</p></div></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><p>Due to the huge workload of counting micronucleus, we tried to develop counting algorithm. We use the algorithm framework of Yolov5 to implement what is available on the author's Github or Model section, which is not covered here. We selected the typical normal nuclei and Micronuclei from the frame of the microscope. There were about 200 chromatin images and 50 micronuclei images in the normal nuclei. We trained a recognition model by using these images as training set. With this model, a large number of imaging results can be analyzed in a short time. We use this model to count micronuclei and measure the cytotoxicity of various substances. The results show that this model is in good agreement with the commonly used counting standards under the experimental conditions. It can be used to compare the difference in toxicity between different substances. For detailed information, please refer to <a href="https://2021.igem.org/Team:HUST-China/Results/Model">Model</a> part. #Color stability testing We would expose our hair dyed with our dye paste and two chemical dyes (red and blue) to constant bright light, take pictures before and after lighting, and calculate the degree of discoloration. The result shows that the light stability of our dye paste is slightly less than that of the chemical dye paste, but the difference is invisible to the naked eye in the actual application scene, so the color of our dye paste can exist stably in the natural light, it's still very practical.</p><div class="image"><img alt="Light exposure's influence on dying" src="https://static.igem.org/mediawiki/2021/f/ff/T--HUST-China--img--result--result-light_exposure%27s_influence_on_dying_.png" style="width: 100%"/><p>Figure 60: Light exposure's influence on dying</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result7" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result7Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result7" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result7Label">result7</h5></div><div class="modal-body"><p>What we did: We cut a bunch of hair in half, pasted half of it onto a piece of cardboard, and exposed it to a constant, bright light for 11 days. Our lights are LED lights and sodium lamps, cycle for 22 hours of light and 2 hours of darkness alternately. At the end of the illumination, half of the hair without illumination was pasted on, and the fade index was analyzed by the algorithm. Considering the change of RGB values, it is difficult to determine the model parameters, and there may be multi-value based on the prediction results given by the neural network. We choose the linear combination of RGB to calculate the gray value, and take the gray value as the index of the concentration. It can be used as an index to evaluate the degree of fading by calculating the change of gray difference between the two groups. This indicator is not linear and therefore should not be understood as an indicator in a percentage sense. On a scale of one to one, the higher the score, the deeper the discoloration. There was no significant difference between indigo and chemical blue, the scores of indigo were 0.178 and 0.161, respectively. Chemical red faded more slowly than lycopene, with a score of 0.186 and 0.313, respectively.</p><p>The process of exploration: When we first carried out the strong light experiment, we irradiated single hair and only treated it for 2 days. The results showed that the dyeing of each hair in the same strand was very different due to the uneven dyeing, and the treatment time is so short that almost all the hair remains the same. In the second experiment, we took a photo of the hair in front of the light and compared it with the photo after the light exposure. The light exposure was extended to 7 days. Although the discoloration was detectable this time, it varied widely between the groups, suggesting that one hair's discoloration was highly accidental. So on the third occasion, we decided to reduce the chance by increasing the amount of hair, using a single strand of hair, and extending the light again to get the difference.</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Color matching experiment and Sumu experiment</h1><p>By adding color aids, mixing two dyes or secondary coloring, we get more hair colors. Now, we have different brightness of the rainbow seven colors and gray, red, brown, tawny and other colors. At the same time, we also used hematoxylin as a dye to explore a series of dyeing. Hematoxylin itself can be brown, add alum can be red, add alkali can be purple. Hematoxylin 's synthetic biology production will be a future extension project.</p><div class="image"><img alt="Olympic rings and Cartoon kid" src="https://static.igem.org/mediawiki/2021/0/04/T--HUST-China--img--result--result-Olympic_rings_and_Cartoon_kid.png" style="width: 100%"/><p>Figure 61: Olympic rings and Cartoon kid</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result8" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result8Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result8" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result8Label">result8</h5></div><div class="modal-body"><p>A coloring list of the three pigments</p><table><thead><tr><th>Picture</th><th>Color</th><th>Coloring method</th></tr></thead><tbody><tr><td><img src="https://static.igem.org/mediawiki/2021/d/d9/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_red.png"/></td><td>Red</td><td>8 ° hair in the room temperature treated with lycopene for 30 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/e/e9/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_orange.png"/></td><td>Orange</td><td>9 ° hair in the room temperature treated with curcumin for 30 minutes then treated with lycopene for another 30 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/6/67/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_light_orange.png"/></td><td>Light orange</td><td>9 ° hair in the room temperature treated with curcumin mixed with lycopene (2:1) for 30 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/1/17/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_yellow.png"/></td><td>Yellow</td><td>9 ° hair in 50 ℃ treated with curcumin for 30 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/f/fc/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_green.png"/></td><td>Green</td><td>7 ° hair in the room temperature treated with indigo for 2 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/2/29/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_cyan.png"/></td><td>Cyan</td><td>9 ° hair in the room temperature treated with indigo for 30 seconds</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/8/89/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_blue.png"/></td><td>Blue</td><td>9 ° hair in the room temperature treated with indigo for 2 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/7/78/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_purple.png"/></td><td>Purple</td><td>9 ° hair in the room temperature treated with indigo for 5 minutes then treated with lycopene for another 10 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/0/0b/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_grey.png"/></td><td>Gray</td><td>9 ° hair in the room temperature treated with lycopene for 5 minutes then treated with indigo for another 30 seconds</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/5/5b/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_reddish_brown.png"/></td><td>Reddish brown</td><td>6 ° hair in the room temperature treated with lycopene for 10 minutes</td></tr><tr><td><img src="https://static.igem.org/mediawiki/2021/8/8b/T--HUST-China--img--result--result-the_teaching_of_dying_exmples_tan.png"/></td><td>Tan</td><td>9 ° hair in the room temperature added tannic acid treated with curcumin for 10 minutes</td></tr></tbody></table></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><div class="image"><img alt="the coloring effect of hematoxylin under various conditions" src="https://static.igem.org/mediawiki/2021/7/7b/T--HUST-China--img--result--result-the_overview_of_all_possible_colors.png" style="width: 100%"/><p>Figure 62: the coloring effect of hematoxylin under various conditions</p></div><h1>Experiment of short peptide perm</h1><h2>Short peptides perming hair experiment</h2><div class="image"><img alt="comparison of short peptides and water" src="https://static.igem.org/mediawiki/2021/6/61/T--HUST-China--img--result--result-comparison_of_short_peptides_and_water.png" style="width: 100%"/><p>Figure 63: comparison of short peptides and water</p></div><p>We used 0.1% and 0.01% short peptides for perm experiment, and set chemical perm and up water perm as control group. The perming effect of short peptide is more obvious than that of ultra-pure water, which shows that in alkaline and 50° conditions, the hair cuticle is opened and the longer react time it takes, the better perming result we get. And after a series experiment, we found that hair dealt with SDS can have more obvious curl effect.</p><div class="image"><img alt="comparison of short peptides with different mixture with other chemicals" src="https://static.igem.org/mediawiki/2021/7/78/T--HUST-China--img--result--result-comparison_of_short_peptides_with_different_mixture_with_other_chemicals.png" style="width: 100%"/><p>Figure 64: comparison of short peptides with different mixture with other chemicals</p></div><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result9" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result9Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result9" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result9Label">result9</h5></div><div class="modal-body"><table><thead><tr><th style="text-align:center">Chemical perm agent(bought online)</th><th>Ultra-pure water</th><th>0.001% short peptide solution</th><th>0.1% short peptide solution</th></tr></thead><tbody><tr><td colspan="4" style="text-align:center">500ul</td></tr><tr><td colspan="4" style="text-align:center">Immerse hair(washed and dried) 20min</td></tr></tbody></table><p>The experimental results showed that the Chemical perm agent had the most obvious effect, while others didn't have any obvious perming effect. We've considered 3 reasons why short peptides have no obvious effect:</p><p>(1) Maybe the peptides didn't get into hair cuticle</p><p>(2) The time for short peptides to form disulfide bonds wasn't enough</p><p>(3) The reaction temperature is not enough.</p><p>So, we get another exploring experiment.</p><p>Time and Temperature:</p><p>(1)Drop a drop of 3M NaOH into 0.01% short peptides solution. [PH test paper: PH=10]</p><p>(2)Control experiment: drop a drop of 3M NaOH into UP water</p><p>(3)Both react at 50° for 1h.</p><p>Re-explore a better condition for short peptides'perming hair</p><p>1)immerse 10 hairs in SDS 3 times to remove charges on hair surface.</p><p>2)Wash and put them in 0.01% short peptides solution</p><p>3)Control experiment: Up water, the subsequent processing steps are the same as 1)2)</p><p>4)both:50°, 1h</p></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><h1>Perming hair stress experiment-9</h1><p>We used different kinds of perms for hairs with 2 groups of different hair, and the hair strains will be damaged after perming. So we designed and conducted stress experiments to test the mechanics of the hair after perming. The figure below shows the experiment data, by which we can know that perming by short peptides will have very little damage to the hair, and short peptide perm damage is less than chemical perm. Moreover, the results of the damaged hair groups showed that the damage of short peptide perm is much smaller than that of chemical perm. Hence, we speculated that perming procedure would cause some damage to hair, but the use of short peptide perm could be used to repair the damaged hair.</p><h3>Healthy hair stress test results (Horizontal: the deformation length; Ordinate: pull force</h3><div class="image"><img alt="Healthy hair stress test results (Horizontal: the deformation length; Ordinate: pull force)" src="https://static.igem.org/mediawiki/2021/4/4b/T--HUST-China--img--result--result-healthy_hair_stress_test_result.png" style="width: 100%"/><p>Figure 65: Healthy hair stress test results (Horizontal: the deformation length; Ordinate: pull force)</p></div><p>It is shown that the hair of the short peptide perm group changes in shape under constantly increasing pull force, and most hair strains can withstand more pull than hair permed by chemical perms. This suggests that short peptide perms do less damage to hair performance than chemical perms. According to the experimental results, we judge that the concentration of short peptides has little effect on the results.</p><h3>Damaged hair stress test results (Horizontal: the maximum length; Ordinate: pull force)</h3><div class="image"><img alt="Damaged hair stress test results (Horizontal: the maximum length; Ordinate: pull force" src="https://static.igem.org/mediawiki/2021/e/e9/T--HUST-China--img--result--result-damaged_hair_stress_test_result.png" style="width: 100%"/><p>Figure 66: Damaged hair stress test results (Horizontal: the maximum length; Ordinate: pull force</p></div><p>This set of data comes from a pulling test of damaged hair. It is shown in this figure that both sets of hairs that use short peptides for perm can withstand more pull than the hairs after chemical perm. And there isn't much difference between the results of short peptide group and that of the water group. We speculate that short peptides can by some means repair the damaged hair, which also portrays the excellent performance of short peptide perm that it can protect the hair and maintain the hair quality.</p><h1>Microscopic effects of PepACS on hair</h1><p>We used scanning electron microscopy to look at healthy hair, bleached hair and bleached hair treated with the PepACS. The results showed that compared with normal hair, the surface of bleached hair was obviously damaged, but no obvious damage was observed after treatment with PepACS, indicating that PepACS may have a certain repair effect, which can be further studied in the future.</p><div class="modal-btn"><button class="btn btn-warning pull-right" data-target="#result10" data-toggle="modal" type="button">Read More</button><div aria-hidden="true" aria-labelledby="result10Label" class="modal fade" data-backdrop="static" data-keyboard="false" id="result10" tabindex="-1"><div class="modal-dialog modal-dialog-scrollable modal-dialog-centered modal-xl"><div class="modal-content"><div class="modal-header"><h5 class="modal-title" id="result10Label">result10</h5></div><div class="modal-body"><p>We soaked the damaged hair with 0.1% PepACS solution and reacted at 50℃ for 3 hours to obtain the damaged hair samples treated with PepACS. No surface damage was clearly observed under scanning electron microscopy after the PepACS treatment, but given the differences between the same batch of damaged hair, we cannot say that the PepACS has a repairing effect. We expect to continue to carry out relevant experiments in the future.</p><div class="image"><img alt="From left to right: healthy hair,bleached hair,bleached hair treated with the PepACS" src="https://static.igem.org/mediawiki/2021/4/4b/T--HUST-China--img--result--result-elect.png" style="width: 100%"/><p>Figure 67: From left to right: healthy hair,bleached hair,bleached hair treated with the PepACS</p></div></div><div class="modal-footer"><button class="btn btn-danger" data-dismiss="modal" type="button">Close</button></div></div></div></div></div><div class="row g-0"><div class="col-4 girl"><img src="https://static.igem.org/mediawiki/2021/2/28/T--HUST-China--img--girl.png"/></div><div class="col-5"><div class="dialogbox"><p>Wow! That’s eye-opening! I guess the old saying that “Rome was not build in a day” does make sense. All those results require accumulation, which is recorded in the <a href="https://2021.igem.org/Team:HUST-China/Notebook">notebook</a>.</p></div></div></div></article></div></div></div></main><footer><div class="container"><div class="row"><div class="col-7"><div class="row justify-content-center"><div class="footer-logo"></div></div><p><span>CONTACT US: </span><a href="mailto:iGEMHUSTChina@163.com">iGEMHUSTChina@163.com</a></p><p>Huazhong University of Sci. & Tech., Wuhan, China</p><p>1037# Luoyu Rd, Wuhan, P.R.China 430074</p><p>Copyright © <span>HUST-China </span>iGEM 2021</p></div><div class="col-5"><div class="row"><div class="footer-xmind"></div></div><div class="row"><div class="footer-snapgene"></div></div><div class="row"><div class="footer-NEBI"></div></div></div></div></div></footer><script src="https://2021.igem.org/Template:HUST-China/content-bundleJS?action=raw&ctype=text/javascript"></script><script src="https://2021.igem.org/Template:HUST-China/mathjax-bundleJS?action=raw&ctype=text/javascript"></script></body></html> |