Model
Project Modeling
Modular Cloning is seen as an easy and efficient way of cloning. This is why we decided to implement the MoClo system for Leishmania tarentolae and by this to improve the use of Leishmania as an expression host for proteins, especially for those proteins that need complicated glycosylation patterns. But is MoClo really that efficient? Let’s see!
As an example we use one of the constructs we created by using Modular Cloning: L1_sAP_ RBD_mCerulean_Strep8His.
It contains the signal peptide of the secretion acid phosphatase (sAP), the coding sequence for the receptor binding domain (RBD) of the Sars-CoV-2 spike protein, mCerulean as a fluorophore, and a Strep/octa-histidine tag for purification. The resulting protein should easily be purified from the cell supernatant by affinity chromatography. The fluorophore allows performing binding assays with the ACE2 receptor such that you can visualize an interaction of both proteins.
The following calculations do not reflect reality because the different problems that can occur during scientific work cannot be calculated in advance (as we had to learn during our project). However, since this is the case for MoClo as well as for classical cloning techniques, like the BioBricks assembly, this factor should have a similar impact on both calculations.
The following amounts of materials are used for one PCR + GelEx: 0.5 µL 10 mM dNTPs, 0.25 µL HF-Q5 polymerase, 1.6 g agarose, 1 GelEx.
The following amounts of materials are used for one MoClo-reaction + plasmid prep + test digest: 2 µL ATP, 0.5 µL BbsI (L0)/0.5 µL BsaI (L1), 0.5 µL T4 DNA ligase, 1 plasmid prep, 0.75 µL BstEII, 0.75 µL XhoI, 0.8 g agarose.
The following amounts of materials are used for one BioBrick assembly reaction + plasmid prep + test digest: 1 µL EcoRI-HF, 1 µL XbaI, 1 µL SpeI, 1 µL PstI, 1 plasmid prep, 0.75 µL BstEII, 0.75 µL XhoI, 0.8 g agarose.

The total costs for the BioBrick assembly are higher than for MoClo, especially because of the larger number of enzymes needed for BioBrick assembly. This is due to the larger number of cloning steps needed for the BioBrick assembly in comparison to the MoClo assembly of the sAP_RBD_mCerulean_Strep8His example construct.

You need seven days to finish a L1-construct with MoClo and twelve days to finish the same construct with BioBrick assembly. The main reason for that are the four cloning reactions and the following transformations and inoculations you need for BioBrick assembly. In comparison, you only need two cloning reactions with following transformations and inoculations for MoClo.
Overall, the calculations show that MoClo is way more cost- and time-efficient than the BioBrick assembly. Especially when you are assembling a whole bunch of constructs as complex as the sAP_RBD_mCerulean_Strep8His, you will need way less time and money with the MoClo system.
To see if reality confirms this theory, we asked our instructors to send us their number of published articles with MoClo in combination with their summed impact factor.
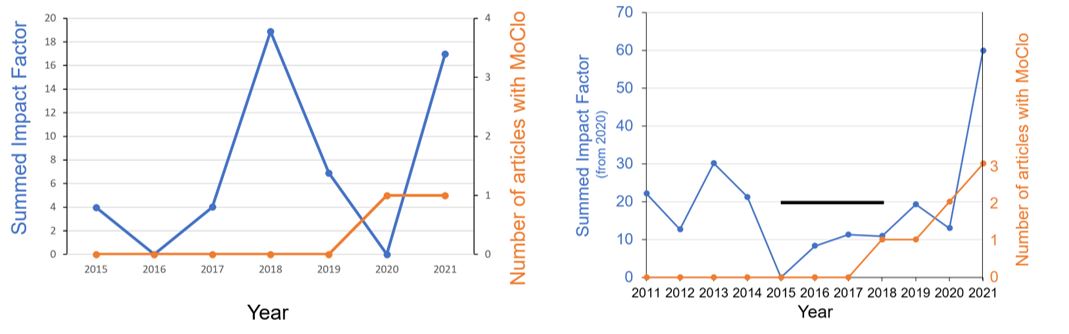
The left graph shows the statistics of Assistant Professor Felix Willmund. When he started to publish articles using MoClo in 2020, his summed impact factor raised to 17 in 2021. The right graph shows the statistics of Professor Michael Schroda. He started to publish articles using MoClo in 2017. Since then, his summed impact factor raised from 10 to 60 in 2021. These statistics indicate that it might be a good idea to use MoClo!
So, there’s only one more thing for us to say: Use MoClo, it’s freaking awesome!
Sources
- Uppsala2013_BioBrick_Assembly_Manual.pdf (igem.org)
- NucleoSpin Plasmid, Mini Kit für Plasmid DNA, MACHEREY-NAGEL MN | MACHEREY-NAGEL (mn-net.com)
- Wizard® SV Gel and PCR Clean-Up System | PCR Purification | PCR Clean Up (promega.de)
- Agarose Standard, 100 g | Agarosen und Gelbildner | Agarosegelelektrophorese | Elektrophoresereagenzien | Elektrophorese | Applikationen | Carl Roth - Deutschland
- https://international.neb.com/products/r3101-ecori-hf#Product%20Information
- https://international.neb.com/products/r0145-xbai
- https://international.neb.com/products/r3133-spei-hf
- https://international.neb.com/products/r3140-psti-hf
- https://international.neb.com/products/m0202-t4-dna-ligase
- https://www.neb.com/products/r3162-bsteii-hf
- https://international.neb.com/products/r0146-xhoi
- https://www.sigmaaldrich.com/DE/de/configurators/tube?product=standard
- https://international.neb.com/products/n0447-deoxynucleotide-dntp-solution-mix#Product%20Information
- https://international.neb.com/products/m0491-q5-high-fidelity-dna-polymerase#Product%20Information
- https://international.neb.com/products/r3539-bbsi-hf
- https://international.neb.com/products/r3733-bsai-hf-v2
- https://international.neb.com/products/p0756-adenosine-5-triphosphate-atp#Product%20Information
- https://armacad.info/blog/highest-impact-factor-journals
