ENGINEERING SUCCESS
Since our project revolved around the creation of a genetic parts library, the design, cloning and testing of basic parts was our daily business. Depending on the genetic source of the respective part, the design process either aimed to create a sequence ready for commercial synthesis or to generate a set of primers that would allow for amplification of the desired sequence from a source plasmid via PCR. In order to demonstrate our engineering success, we want to introduce you to one of the most vital parts of our expression system, the sAP secretion signal.
The sAP signal tag is a secretion signal peptide derived from the secretory protein secreted acid phosphatase 1 (sAP1) which was originally purified from Leishmania mexicana, a human pathogenic Leishmania strain[1]. It mediates the exocytosis of the protein after its biosynthesis in the endoplasmatic reticulum (ER). This is extremely useful for recombinant protein production, since target proteins can be harvested directly from cell culture supernatant without the need for cell lysis. Since post-translational glycosylation of proteins happens within ER and Golgi apparatus, along the secretory pathway [2], the sAP1 signal peptide enables the production of glycosylated proteins in Leishmania. This was a vital aspect to our project, because we aimed to demonstrate Leishmania’s complex, human-like glycosylation capabilities which make them such a promising expression host for biopharmaceutical proteins. During the realisation of this project, the SARS-CoV-2 receptor binding domain (RBD) functioned as our exemplary protein of interest. Not only would transgenic expression of the SARS-CoV-2 RBD prove our expression system’s scientific and medical relevance, it would also shine a light on the post-translational modification capabilities of Leishmania, since the receptor binding domain is a heavily glycosylated protein.
So now that you know how important protein secretion is to the objective of our project, it doesn’t come unexpected that most of the L1 constructs we created with our MocloMania collection contain the N-terminal sAP secretion signal. But how did we design it and construct it?
DESIGN
Well, the sequence to this part was directly derived from the commercial pLEXSY_I-blecherry3 plasmid, distributed by JenaBioscience, which is the basisof our L1 expression vector weird_plex.
pLEXSY_I-blecherry3 is a Leishmania expression vector that was specifically designed to allow for either cytosolic or secretory protein expression. The Leishmania mexicana derived sAP secretion signal peptide is thus already included in the plasmid's backbone. JenaBioscience constructed the cloning frame in a way that would allow for either inclusion or exclusion of the signal peptide from the open reading frame, depending on the choice of restriction enzymes used for target protein insertion[3].
During the domestication of pLEXSY_I-blecherry3 for the MoClo system, we introduced a LacZ-alpha gene, but eliminated the sAP secretion sequence. Instead, the sequence was adapted and turned into a functional L0 basic part, the sAP secretion signal that we are looking at right now. This way, switching between cytosolic and secretory protein expression can be even more easily coordinated, simply by adding or leaving out parts in a MoClo reaction.
The very first step in our endeavour to create this part was to find its sequence within the pLEXSY_I-blecherry3 backbone and double-check it with protein sequence data found in online resources such as UniProt, see Figures 1 and 2.


After confirming the exact position of the sequence within the pLEXSY_I-blecherry3 backbone, we had to retrieve it from the plasmid. For this, we designed a specified set of primers called sAP-forward and sAP-reverse that would allow for amplification of the sAP sequence via PCR, while simultaneously introducing specified four nucleotide overhangs and BbsI restriction sites to the sequence. These additions are what makes the sequence suitable for application within our Modular Cloning system.
For the sAP secretion signal to be functional, it has to be attached to the target protein's very N-terminal region. Thus, we decided to adapt the part to B2, our most upstream cloning position. This meant that the respective overhangs that needed to be attached were CCAT to the 5' end and AATG to the 3' end of our sequence. For more information on primer design as well as overhang choice, feel free to check out our tutorial on how to generate a MoClo part.

We tested our primer design by going through the entire amplification and cloning process in silico. When we found everything to work out well, it was time to order the primers. We also set up an experimental plan for the upcoming weeks.
EXPERIMENTAL PLAN
1 | PCR with ensuing gel electrophoresis and gel extraction
2 | MoClo ligation of extracted fragment into B2 plasmid backbone
3 | Transformation, plasmid prep and test digest to confirm successful insertion
4 | Assembly of resulting L0_sAP_B2 into L1 construct
5 | Transformation, plasmid prep and test digest to confirm successful assembly
6 | Transfection into Leishmania tarentolae cell culture
7 | Investigation of protein expression via immunostaining on Western blot
So upon arrival of the primers, we were ready to go!
CONSTRUCTION
Right after arrival of the primers, we resuspended and diluted them to the desired concentration and then conducted a standard PCR amplification reaction using NEB's Q5 Polymerase. As a template, we used the unprocessed pLEXSY_I-blecherry3 plasmid. After completion of the thermocycling process, we loaded the resulting PCR sample onto a 2% agarose gel, accompanied by the Thermo Fisher LowRange GeneRuler. We did this because thanks to our in silico amplification, we knew that we were expecting a small, 100 bp fragment.

1 | PCR sAP-forward + sAP-reverse on pLEXSY_I-blecherry3 L | Thermofisher LowRange GeneRuler [bp]
We observed the predicted 100 bp fragment on the gel which meant that we could continue with our experimental plan. By extracting the fragment from the agarose gel with the help of a MACHEREY-NAGEL Gel Extraction Kit we recovered the PCR fragment, free from any residual template DNA or enzymes present in the PCR reaction. This allowed us to directly introduce the recovered fragment into a MoClo ligation reaction in order to insert it into pAGM1276, the B2 plasmid backbone adapted from the Weber MoClo plasmid collection [4]. Introduction into a plasmid backbone doesn't only preserve our fragment from degradation, it also helps us to evaluate whether the cloned PCR fragment was amplified accurately and really carries the desired overhangs.
As a means of demonstration, we have included a standard MoClo Ligation protocol that we used standardly when it came to generation of L0 basic parts. For a MoClo reaction to be efficient, it is important to add all genetic components, backbone as well as insert, in equal stoichiometric quantities. For many protocols this is 40 fmol, but other values such as 20 fmol are published as well[5]. Thanks to photometry, we were able to quantify the concentrations of our pAGM1276 as well as of our gel-excised sAP fragment. With the help of the Bioline online molar quantity calculator, this allowed us to predict the exact sample volume that needed to be added to the reaction mix in order to give 40 fmol sample molecules each.
L0 MoClo ligation | 20 µL reaction volume
2 µL | 10x NEB CutSmart-Buffer
2 µL | ATP
0,5 µL | BbsI
0,5 µL | T4 DNA Ligase
40 fmol | sAP Fragment
40 fmol | pAGM1276
fill up | ddH2O
After addition of all reactants (enzymes last), the reaction mix was briefly vortexed, spun down and then put into a thermocycler to undergo the standard MoClo cycling process. Every protocol does this a little differently, but the basic cycle program is always the same.
L0 MoClo ligation | Thermocycling program
45x--------------------------------
37°C | 2 min | 16°C | 5 min
--------------------------------
1x
--------------------------------
37°C | 5 min
80°C | 10 min
16°C | indefinitely
--------------------------------
After completion of the MoClo thermocycling run (usually performed over night), 10 µL of the resulting sample were transformed into competent TOP10 E. coli. This E. coli strain allows for rapid and efficient amplification of the hopefully ligated L0 construct and is also suitable for blue-white screening. Since our goal was to insert the sAP sequence into the B2 backbone pAGM1276, we will proceed to call the cloned construct L0_sAP_B2, according to our standardized Moclo Mania nomenclature.
After transformation of the sample and incubation of the transformed E. coli at 37°C over night on an IPTG/XGal-agar plate, white and blue colonies could be observed the next morning. Because the lacZ-alpha gene fragment in pAGM1276 is excised upon insertion of the sAP fragment, the resulting transformants don't have all the parts it takes to produce functional beta-galactosidase. This means that the white colonies are the ones carrying our successfully cloned construct. We inoculated several of them into liquid culture and let them grow on a shaker at 37°C over night. Both, the LB medium for the agar plates and that for the liquid culture had spectinomycin added, since this is the antibiotic that our L0 plasmid backbones carry a resistance marker against.
The next morning, the liquid cultures showed substantial growth and could be prepped with the help of a MACHEREY-NAGEL DNA MiniPrep Kit. This way, the replicated L0_sAP_B2 construct could be extracted from the E. coli cells and purified. We ended up with our purified, (hopefully) correctly assembled L0 construct in a microfuge tube on ice. Now, in order to verify that the DNA in our tube is really the construct we are looking for and not just some random E. Coli plasmid, we performed a test digest. Again, going through this process in silico was very helpful to us, because it allowed us to easily plan the most feasible digest, rendering the most distinct band pattern and only requiring easily applicable enzymes with high activity in the same buffer. We finally settled on a digest using BstEII as well as XhoI. Doing a double digest like this was particularly handy, because XhoI cuts within the actual sAP sequence. This means that the successfully cloned construct would result in two bands, whereas the empty pAGM1276 backbone could only be linearized.
Test digest | 20 µL reaction volume
2 µL | 10x NEB CutSmart-Buffer
0,75 µL | BstEII
0,75 µL | XhoI
500 ng | DNA sample
fill up | ddH2O
Incubate at 37°C for 1 h.

1 | pAGM1276 | 2842 bp
2 | L0_sAP_B2 | 1705 + 615 bp
L | Thermo Fischer GeneRuler Plus Ladder [bp]
As can be seen in Figure 4, the empty pAGM1276 shows one band, whereas the digested L0_sAP_B2 displays the exact two band pattern that we anticipated from our in silico digest. This is already a good indicator that our L0 part was cloned successfully! But what if point mutations were introduced during PCR, too small to be seen with gel electrophoresis? In order to dispel any doubts and verify the correct insertion and sequence of our sAP secretion peptide once and for all, we sent the plasmid off to commercial Sanger sequencing at MICROSYNTH, a Swiss-based sequencing company with a branch in Germany.
For this, we relied on a set of sequencing primers that have been designed by our supervising working group in order to facilitate sequencing of L0 basic parts. Due to the high similarity between the indivdual L0 plasmid backbones (they only really differ at the insertion site), these primers called L0-Seq-forward and L0-Seq-reverse can be used for sequencing of any and all L0 parts, independently of their cloning position.
L0-Seq-forward | aatacgcaaaccgcctctcc
L0-Seq-reverse | aataggcgtatcacgaggc

Only three days after sending our sample off to sequencing, we got the result! It's a bit like opening a bag of wonders every time you click on those sequencing files. You just never know wether things are going to allign perfectly or wether nothing seems to match at all! Luckily, our L0_sAP_B2 looked very good, as you can see in Figure 5. The sequence of the processed L0-Seq-forward initiated fragment matches that of the in silico cloned L0_sAP_B2 perfectly, except for usual sequencing uncertainties in the very terminal regions of the fragment. This was great news!! Now we could continue our experimental journey by assembling our L0 secretion signal into a variety of L1 constructs. Thanks to the practicability of Modular Cloning, this wasn't a very difficult endeavour.
Now we have already elaborated on the great importance of the sAP secretion signal to the prospect of our project, so it doesn't come as a surprise that as soon as we had the finished L0_sAP_B2 part, we immediately started implementing it into all of our L1 constructs. In order to simplify this demonstration, let's just look at the MoClo ligation reaction performed for generating L1_sAP_RBD_Strep8His. This L1 composite part contains the sAP secretion signal, a coding sequence for the SARS-CoV-2 receptor binding domain and a C-terminal Strep8His purification tag.
You will see that the reaction mix is basically identical to that of the L0 ligation, except now we have to remember the two-enzyme system of MoClo and employ BsaI as our executing type IIS restriction enzyme. Also, instead of a L0 plasmid backbone we now want to include our L1 expression vector, weird_plex, that will allow us to transfect and express our construct within Leishmania tarentolae. Thermocycling conditions for this ligation are virtually identical to the ones for a L0 ligation. The only exception is that, after cycling, a 5 min incubation is performed at 50°C rather than 37°C, because BsaI has a slighlty different activity pattern than BbsI.
L1 MoClo ligation | 20 µL reaction volume
2 µL | 10x NEB CutSmart-Buffer
2 µL | ATP
0,5 µL | BsaI
0,5 µL | T4 DNA Ligase
40 fmol | L0_sAP_B2
40 fmol | L0_RBD_B3_B4
40 fmol | L0_Strep8His_B5
40 fmol | weird_plex
fill up | ddH2O
Again, after completion of the MoClo thermocycling run, 10 µL of the resulting sample were transformed into competent TOP10 E. coli. And yet again, after over-night incubation on IPTG/Xgal agar plates, we inoculated the white colonies into liquid culture. This time, though, both the LB medium for the agar plates as well asand that for the liquid culture had ampicillin added, because weird_plex carries its own E. coli expression cassette that includes an ampicillin resistance marker.
The next morning, the liquid cultures were again prepped with the help of the MACHEREY-NAGEL DNA MiniPrep Kit and the (hopefully) assembled L1_sAP_RBD_Strep8His construct was extracted from the E. coli cells. To verify that our construct was correctly assembled, we performed another test digest. In silico cloning and digestion helped us to choose PstI, because it gives a very nice and distinct band pattern.

1 | L1_sAP_RBD_Strep8His | 3894 + 1323 + 991 + 991 + 757 bp
L | Thermo Fischer GeneRuler Plus Ladder [bp]
As you can see in Figure 6, our test digest on the L1 construct resulted in the expected bands. This means that the plasmid in our tube is of the predicted size, which is a very good sign, since this probably means that all L0 parts have been assembled into the L1 backbone successfully. To rule out any assembly errors and to verify the identity of our prepped L1 construct with 100% certainty, we again sent the plasmid off to Sanger sequencing at Microsynth.

Yayy! After three days, the sequencing results are back and the alignment looks super sharp! Except from a few bases that were probably missed due to clustering of identical bases (Sanger sequencing always struggles with repetitive single nucleotide sequences), everything looks super promising. This means that we not only successfully constructed our basic L0_sAP_B2 part, but we have also managed to assemble it into a complete L1 construct. Now, as mentioned before, we of course didn't only do this for L1_sAP_RBD_Strep8His, but for many other constructs that are all ready for the next experimental round. Transfection into Leishmania!
EXPRESSION
Now that we have mastered all the cloning work, it is time to move on to the second big step in our project - expressing the genetic constructs in Leishmania tarentolae. To do this, we of course have to somehow get our construct into Leishmania cell first. How do we do this? By transfecting the cells via electroporation. This procedure as well as all other labwork involving the cultivation of Leishmania is based on the protocols provided by our second supervising working group as well as the protocols distributed by JenaBioscience in their commercial LEXSinduce expression kit. If you are interested in finding out more about the specifics of working with Leishmania, please feel free to check out our guide on how to handle Leishmania!
After transfecting the parasites with our L1_sAP_RBD_Strep8His, the Leishmania are brought out onto agar plates. Here, addition of the antibiotic bleomycin ensures that all growing colonies must have taken up our plasmid, since the L1 expression vector weird_plex is equipped with a bleomycin resistance gene. It takes anything from 5 to 9 days for colonies of sufficient size to form on the plate. These can then be inoculated into liquid medium where the parasites can be grown and re-diluted into ever larger volumes. Once the cell culture has reached the desired density, protein expression can be induced by addition of tetracycline to the culture medium. For more in-depth information on the specific features of the weird_plex expression vector and its inducible expression, feel free to head over to the weird_plex part page.
Tetracycline induction is held stable for around 2-3 days, until the cell culture is finally harvested. By spinning down the culture in a centrifuge for over an hour, the cells are separated from the culture medium and form a pellet. Depending on the localisation of the expressed protein, this pellet can be lysed and the resulting lysate either purified (cytosolic expression) or cooked up with SDS sample buffer for analysis (secreted expression). In our case most of the L1 constructs, including L1_sAP_RBD_Strep8His, mediate secretion of the recombinantly expressed protein into the culture medium. Thus, after centrifugation, the cell culture supernatant is carefully collected in order to immediately undergo downstream purification procedures. Only a small sample is saved for analysis purposes. The sample is treated with trichloroaceticic acid in order to precipitate all protein and the resulting pellet cooked up with SDS sample buffer. Along with the cell lysate sample as well as control samples of untransfected Leishmania cell culture, it is loaded onto an SDS-gel for SDS-PAGE. After western blotting, the samples can be immunostained with the help of different antibodies in order to visualize the target protein. In the case of L1_sAP_RBD_Strep8His, we chose antibodies against RBD to stain against our fusion protein.
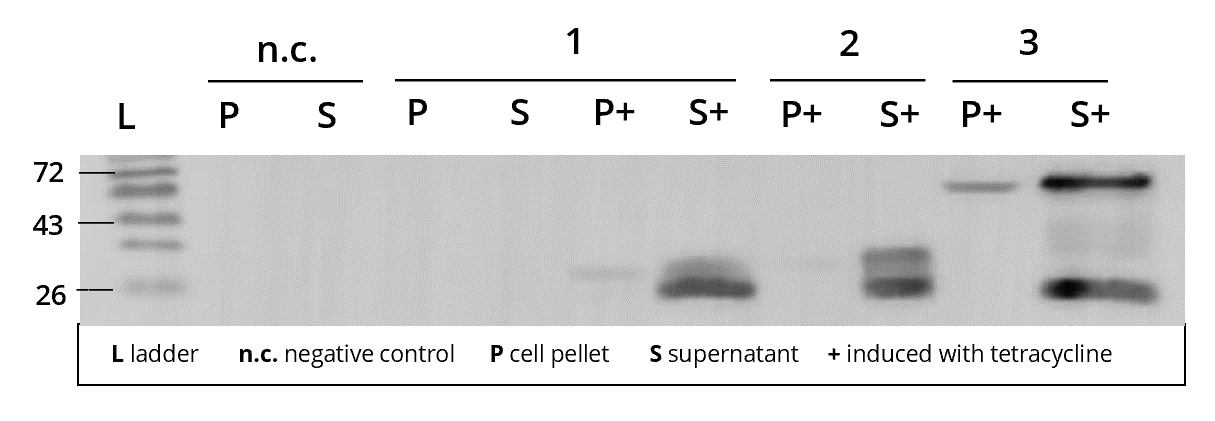
1. AB | ms anti-RBD
2. AB | rb anti-ms HRP
1 | L1_sAP_RBD_Strep8His | 27.7 kDa
2 | L1_sAP_Myc_RBD_Strep8His | 30.2 kDA
3 | L1_sAP_RBD_GST | 51.9 kDa
n.c. | negative control | untransfected Leishmania culture P cell pellet | S cell culture supernatant | + culture induced with tetracycline |
L | Thermo Fischer PageRuler Protein Ladder [kDa]

1. AB | ms anti-RBD
2. AB | rb anti-ms HRP
1 | L1_sAP_RBD_mVenus | 52.1 kDa
2 | L1_sAP_RBD_mVenus_GST | 78.8 kDA
3 | L1_3xHA_RBD_mVenus | 55.9 kDa
4 | L1_3xHA_RBD_HA8His | 28 kDa
p.c. | positive control | RBD-GFP fusion protein | 54 kDa
n.c. | negative control | Leishmania culture transfected with empty weird_plex
P cell pellet | S cell culture supernatant | + culture induced with tetracycline |
L | Thermo Fischer PageRuler Protein Ladder [kDa]
After loading several different cell cultures transfected with our secreted fusion proteins onto a blot and staining against the SARS-CoV-2 receptor binding domain, see Figure 8, we detected a significant expression of our recombinant protein in all three transfected cell lines, 1-3! As expected, the negative control n.c., a Leishmania culture that wasn't transfected with any L1 expression vector, showed no bands. Furthermore, protein expression could only be detected in cultures that were induced with tetracycline, P+/S+, advocating the efficiency of the tet-controlled transcriptional activation. Looking back at our sAP secretion tag, we can observe that bands of the expected size appear in both the cell lysate P as well as the culture supernatant S, but signal intensity is much higher in the supernatant for all three cultures. The faint bands in the lysate samples indicate that a small part of our fusion protein might still be on the way out of the cells. Yet, the vast majority of protein is successfully secreted into the culture medium! The same holds true for cultures 1 and 2 in Figure 9 where protein expression was only detected in the cell culture supernatant. This proves the functionality of our sAP secretion signal and highlights its importance in producing complexly glycosylated protein that need to pass the secretory pathway.
PURIFICATION
After expression in Leishmania, we of course also wanted to harvest the fruit of our work, our secreted recombinant protein. The best way to do this is to incorporate a purification tag into the fusion protein, ideally allowing us to purify it directly from crude cell culture supernatant with the help of gravity flow/batch purification procedures. These procedures certainly hold the most room for improvement when it comes to the ongoing implementation of our project, since many purification attempts have remained unsuccessful. We are still working hard on figuring out what factors can be altered in order to increase purification yield. Since Strep-purification has not given good results yet, we cannot continue our demonstration by following L1_sAP_RBD_Strep8His all the way down the line. Instead we'll take a look at a GST-tagged fusion protein, L1_sAP_RBD_GST, that shows the most promising results and gives further evidence to the quality of our sAP secretion signal!

culture was transfected with L1_sAP_RBD_GST and protein expression induced with tetracycline
blot was co-stained against both RBD and GST a) Culture supernatant b)Lysate after cell debris
There are two things that we can conclude from the blots in Figure 10. First, there is an issue with our GST-tag being cleaved off prematurely, rendering a lot of singular GST and decreasing the purification yield of our RBD_GST fusion protein. Second, there are differences between the samples purified from the cell lysate (a) and the ones purified from the culture medium supernatant (b). We can see that the intensity of the immunostain is much higher in the supernatant purification, suggesting that recombinant protein quantities in the culture medium exceeded those of the cell lysate by far. Thus, our sAP secretion tag effectively mediates translocation of the target protein into the cell's exterior!
Finally, we could not only show that the sAP secretion peptide has been successfully adapted as a MoClo basic part, which can be assembled into cohesive L1 constructs, we were also able to show that it can be transgenically expressed in Leishmania tarentolae and mediates exocytosis of the recombinant target protein into the surrounding cell culture medium.
Now of course, most of our cloning, expression and purification efforts didn't show complete success upon first trial, so keep in mind that for many of our parts, including L0_sAP_B2, the generation process was a lot more tedious than the idealized description given above. By tweaking parameters such as thermocycling settings during PCR or DNA concentrations during ligation, most of these obstacles were eventually overcome and led us to build the extensive part collection that we are proud to present to you today! Feel free to check out our lab journal to catch a first-hand view on our experimental approaches. And as always, reach out to us if you have any further questions!
Sources
- Ilg T, Stierhof YD, Etges R, Adrian M, Harbecke D, Overath P. Secreted acid phosphatase of Leishmania mexicana: a filamentous phosphoglycoprotein polymer. Proc Natl Acad Sci U S A. 1991;88(19):8774-8778. doi:10.1073/pnas.88.19.8774
- Schjoldager, K.T., Narimatsu, Y., Joshi, H.J. et al. Global view of human protein glycosylation pathways and functions. Nat Rev Mol Cell Biol 21, 729–749 (2020). https://doi.org/10.1038/s41580-020-00294-x
- https://www.jenabioscience.com/files/jenabioscience/datasheet_extern/EGE-1410.pdf, pages 9, 18, last visited 10/17/21, 11:00 ECT
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S (2011) A Modular Cloning System for Standardized Assembly of Multigene Constructs. PLoS ONE 6(2): e16765. https://doi.org/10.1371/journal.pone.0016765
- Marillonnet, S., & Grützner, R. (2020). Synthetic DNA assembly using golden gate cloning and the hierarchical modular cloning pipeline. Current Protocols in Molecular Biology, 130, e115. doi: 10.1002/cpmb.115
