...GENERATE A MOCLO PART
HOW TO GENERATE A GENETIC PART FOR MODULAR CLONING IN LEISHMANIA
3xHA detection tag? Great! mVenus fluorescent tag? Amazing!
But how do you make your gene of interest suitable for Modular Cloning in Leishmania, so that you can benefit from the purification and detection tags provided in our kit?
Well, it’s a lot simpler that one might think. Let’s walk together through the steps of designing, synthesizing and cloning an original L0 part with your gene of interest for expression in Leishmana tarentolae. For demonstration purposes, we’ll pick another exemplary protein. The human glycoprotein erythropoetin (short EPO) seems to be a suitable candidate. It’s a signaling protein produced by fibroblasts in the kidney and induces red blood cell production in the bone marrow. Recombinant EPO is an important biopharmaceutical for the treatment of anaemia, a shortage of red blood cells that often occurs in patients with chronic kidney failure or as a side effect of cancer treatment.
So now that we have picked our example protein, our objective is to generate a basic genetic part that is suitable for Modular Cloning and can thus be equipped with a purification tag from our MoClo Mania collection. Furthermore, the part has to be functionally expressed in Leishmania tarentolae. Let’s get started!
1. GETTING THE SEQUENCE
The very first step to generating your own basic part is, of course, to get your hands on the sequence that encodes your protein of interest. Do you already have access to genetic material, say the EPO gene within a plasmid vector? In this case, you have to be aware that adequate codon usage can be crucial for functional target gene expression in the recombinant expression host. [1] Since your gene source might be tailored towards the codon usage bias of another organism, e.g. E. coli, you must consider having your gene of interest newly synthesized for expression in Leishmania. If the differences in codon usage appear marginal, e.g. in rather short genetic sequences, a simple PCR might be enough to make your gene suitable for Modular Cloning. You can find more information on this in Step 5.
We choose the best possible way, i.e., synthesizing the gene encoding EPO with optimal codon usage for Leishmania tarentolae.
In order to retrieve the amino acid sequence of the human EPO protein, we browsed the NCBI gene databaseand, with a little bit of digging, found the entry easily (NCBI gene ID: 2056 -> NCBI reference sequences -> consensus CDS: CCDS5705.1). A quick check in the UniProt database (UniProtKB - P01588 (EPO_HUMAN) tells us about the amino acid sequence of the human erythropoeitin precursor. It also lets us know that the first 27 amino acids are part of a signal peptide and that there are multiple sites of N-linked as well as O-linked glycosylation.

MGVHECPAWLWLLLSLLSLPLGLPVLGAPPRLICDSRVLERYLLEAKEAENITTGCAEHCSLNENITVPD TKVNFYAWKRMEVGQQAVEVWQGLALLSEAVLRGQALLVNSSQPWEPLQLHVDKAVSGLRSLTTLLRALG AQKEAISPPDAASAAPLRTITADTFRKLFRVYSNFLRGKLKLYTGEACRTGDR
Now that we have our desired amino acid sequence, we can move on to the second step.
2. OPTIMIZING CODON USAGE
Thanks to the work of many smart bioinformaticians, handling the different molecular states of a piece of biological information has never been easier. A variety of online tools allows for easy manipulation and interconversion of DNA, RNA and protein sequences at will. One of these services is particularly useful when it comes to codon optimization. The Reverse Translate tool of the Sequence Manipulation Suite (SMS) website generates DNA sequences based on a protein sequence input, all the while employing a codon usage bias of choice. As mentioned before, the codon usage bias may critically affect an organism’s capability to express recombinant genetic material. So in order to make our EPO suitable for expression in Leishmania tarentolae, we’ll have to reverse translate our amino acid sequence while employing Leishmania tarentolae codon usage. In reality, this is just a few clicks of work. The following tutorial will give you a quick overview as to how it might look on your screen:

Visit this websiteand paste your desired amino acid sequence into the upper input field. Next, the tool needs a codon usage table that it can base its reverse translation on. You can either just use the one that we provide here or you can find it for yourself by clicking the link on the website forwarding you to the Codon Usage Database.
Here you will find a QUERY box in which you can simply search for Leishmania tarentolae and then pick the whole genome codon usage. Now you should be seeing something like this:
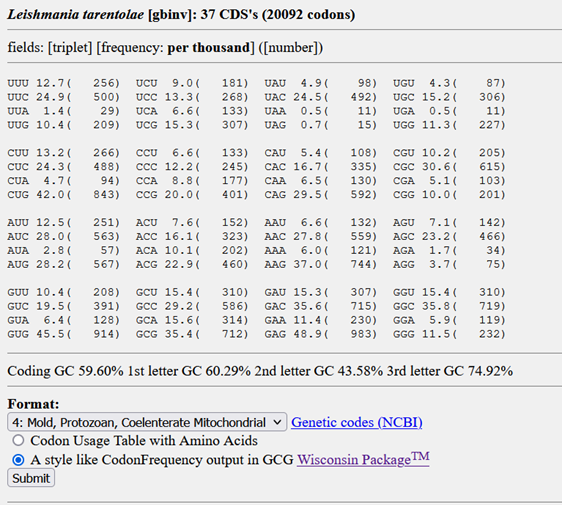
While this is clearly the right table, it is not in the right format yet. The Reverse Translate tool requires codon usage tables to obey the GCG format. To achieve this, you must choose a genetic code which, in our case, is that of a protozoan, so pick Nr. 4. Then make sure to highlight the CodonFrequency output in GCG Wisconsin Package™, click submit and you’re good to go. By now, you should have ended up with a table looking like this:

This is exactly what we are looking for! So just go ahead, copy all of it (including the header row) and paste it back into the lower input field of the Reverse Translate page. Now the time has finally come for some good old reverse translation. Simply hit the submit button and see:

You should be faced with something like this. This little pop-up contains a ton of information, but only the very first paragraph is of interest to us, the one showing the reverse translation to a sequence of most likely codons. You can simply copy this sequence and paste it somewhere safe and boom– you have your genetic sequence optimized for Leishmania tarentolae.
ATG GGC GTG CAC GAG TGC CCG GCG TGG CTG TGG CTG CTG CTG AGC CTG CTG AGC CTG CCG CTG GGC CTG CCG GTG CTG GGC GCG CCG CCG CGC CTG ATC TGC GAC AGC CGC GTG CTG GAG CGC TAC CTG CTG GAG GCG AAG GAG GCG GAG AAC ATC ACG ACG GGC TGC GCG GAG CAC TGC AGC CTG AAC GAG AAC ATC ACG GTG CCG GAC ACG AAG GTG AAC TTC TAC GCG TGG AAG CGC ATG GAG GTG GGC CAG CAG GCG GTG GAG GTG TGG CAG GGC CTG GCG CTG CTG AGC GAG GCG GTG CTG CGC GGC CAG GCG CTG CTG GTG AAC AGC AGC CAG CCG TGG GAG CCG CTG CAG CTG CAC GTG GAC AAG GCG GTG AGC GGC CTG CGC AGC CTG ACG ACG CTG CTG CGC GCG CTG GGC GCG CAG AAG GAG GCG ATC AGC CCG CCG GAC GCG GCG AGC GCG GCG CCG CTG CGC ACG ATC ACG GCG GAC ACG TTC CGC AAG CTG TTC CGC GTG TAC AGC AAC TTC CTG CGC GGC AAG CTG AAG CTG TAC ACG GGC GAG GCG TGC CGC ACG GGC GAC CGC
3. CHECKING FOR RECOGNITION SITES
Now you think you might be ready for finally getting down to business, huh? WRONG! There is one more essential step missing. We now made sure that our genetic sequence matches Leishmania’s requirements for adequate gene expression. But does it match the criteria necessary for usage in Modular Cloning reactions?
While MoClo certainly comes with a lot of advantages - mainly efficiency and its modularity - it also comes with a downside. The genetic parts employed within a MoClo reaction must always be free of any intrinsic BsaI/BbsI recognition sites. These two type II S restriction enzymes are the enzymatic foundation to the entire Modular Cloning method and it is of utmost importance to eliminate any „wild" recognition sites within our gene of interest, because otherwise our genetic part might just be cut into pieces once we try to actually make use of it. We certainly do not want that to happen, so let’s make sure to employ our software of choice in order to check for any endogenous BsaI/BbsI recognition sites within our optimized DNA sequence.

Did you find any?
NOPE. Well, aren’t you lucky! As you can see in the image above, our erythropoeitin contained a lot of recognition sites, but none for BsaI or BbsI.
YESSS. Now that’s a bummer! Due to the codon optimization, intrinsic BsaI/BbsI restriction sites should be rather rare. If you still happen to stumble upon one, don’t fret! After all, you’re having the part synthesized which means that you have total control over the nucleotide make-up of its sequence. Take a closer look at the intrinsic recognition site and try to introduce silent mutations that will disrupt the recognition motif. Meanwhile, always keep an eye on the codon usage table to ensure that you’re not messing with the amino acid sequence itself.

Let’s say your gene of interest contains the sequence GAGAAGACG. An intrinsic BbsI site that nobody saw coming! A quick peak at Leishmania tarentolae‘s codon usage table will tell you that GAG codes for the amino acid glutamic acid, appearing around 49 times among 1000 codons of the Leishmania transcriptome. The triplet GAA also codes for glutamic acid, even though it only appears a mere 11 times among 1000 codons. But introducing a codon that‘s a little less favorable is a much better trade-off than having your genetic part cut into shreds by the restriction enzymes. So let’s just switch GAG to GAA and we’re good to go! We can proceed with step Nr.4.
4. CHOOSING A CLONING POSITION
Now it’s time for some hands-on MoClo action. Have you thought about what cloning position you want to put your gene of interest into? It really depends on your indivdual experimental needs and on the pre-existing parts that you might be utilizing from our MoClo Mania collection.
For erythropoeitin, we know that the protein is subject to multiple glycosylations, so we’ll want to employ the sAP-tag secretion peptide that will steer our protein into the secretory pathway and make sure that it gets all the glycosylation action it needs. Beyond that, a purification tag such as the Strep8His-tag would be cool because it will allow for easy purification of our EPO from the cell culture supernatant. Checking back with our MoClo Mania collection, the cloning position B3_B4 seems to be a reasonable choice for our EPO part. Knowing this, we can simply take a look at the overhang table (at the very top of this wiki page) and see that we’ll need to add AATG to the 5‘ end of the EPO gene and TTCG to its 3‘ end. Confused as to where these funky four nucleotide sequences are coming from? Check out our page on Modular Cloning for more detailed explanations!

Once you’ve chosen your desired cloning position and taken note of the respective overhangs, we can move on to step Nr. 5.
5. ADD BBSI RESTRICTION SITES
Wait – in step 3 we were told to eliminate all possible BbsI restriction sites and now we’re supposed to add new ones back in? I promise there is method to this madness! The new BbsI restriction sites will be added on the very ends of our gene of interest, along with the respective 4 nucleotide overhangs. Why BbsI, you’re asking? Check our page on Modular Cloning for more information. Basically, the restriction sites will allow us to assemble the gene part into a L0 plasmid backbone. This will not only make sure that the part is actually equipped with the right overhangs, it will also allow for its storage in a stable form over long periods of time, e.g. in your very own MoClo library.
BbsI has the recognition site GAAGAC NN XXXX, with NN being two arbitrary bases and XXXX being the four-nucleotide overhang that is left after cleavage. We need this overhang to be exactly the overhang that is attributed to the MoClo cloning position the part is supposed to fill. This is vital, because it allows our part to be standardized and ready for its assembly with other basic parts in the correct, pre-determined order. For starters, we can help ourselves by highlighting the very first and last base triplet of our sequence of interest. Now when it comes to the addition of the overhangs, let’s look at both of them individually:
A. B2/B3 – AATG
On the 5‘ end of our EPO, we need to have the overhang AATG. Looking at the genetic sequence, we can see that the very first base triplet is already an ATG, coding for the amino acid methionine. This
is very helpful because all we need to do is add an additional A in front of the sequence and we’ve already got our overhang down. Now all there is left to do is fill in two random nucleotides and then introduce the BbsI recognition site.
Going off the leading strand, you can remember that the 5‘ end will always carry the recognition site as GAAGAC NN XXXX.

B. B4/B5 – TTCG
We’ve decided for our EPO part to stretch across cloning positions B3 as well as B4, meaning that the next overhang we have to consider is the one inbetween B4 and B5, namely TTCG. Now things
are a little different here. We can’t just attach TTCG to the 3‘ end of our gene of interest, because this would mess up the downstream reading frame pretty badly. In the overhang table, the underline within the overhang represents
the codon that is read in-frame. It says TTCG, meaning that TCG ought to be in-frame with our gene of interest. In order to guarantee this, we have to squeeze two more nucleotides inbetween the 3‘ end of our
gene and the beginning of the overhang. Together, with the initial T of the overhang, they have to form a functional codon. And ideally, this codon translates to a small residue amino acid that will not mess with the higher structures of our resulting
fusion protein. A quick look at Leishmania tarentolae’s codon usage table reveals that GGT, coding for glycine, should be a viable option. So to the 3‘ end of our EPO, we’ll add GGTTCG, followed by two arbitrary
nculeotides and then the BbsI recognition site. Going off the leading strand, you can remember that the 3‘ end will always carry the recognition site as XXXX NN GTCTTC.
Additionally to your recognition site, it is important to stretch the genetic sequence a little beyond the limits of the recognition sites, since BbsI is an endo-, not an exonuclease. You can pretty much add whatever nucleotide sequence you find most useful, but since you are having your part synthesized, you might want to add a standard sequence against which you have suitable primers at hand. This will allow you to amplify your synthesized DNA fragment with a simple PCR. For our EPO, we added a poly-A-tail just to function as an arbitrary place-holder.
If you reverse translated your protein sequence, then everything should be fine, but just to double-check - please make sure that there are NO STOP CODONS left at the 3‘ end of your part! This would be detrimental to the successful expression of your fusion protein. If you, by any chance, happen to be designing a L0 part for cloning position B5, then you must forget what I just said and make sure that you DO HAVE a stop codon. B5 is the most downstream part of the resulting transcriptional unit and thus needs to be equipped with a translation terminating sequence.
Now we have added our overhangs and recognition sites and are left with a linear DNA sequence that we’ll call EPO-B3_B4. You’ve done the same? Great! Then there‘s only one more step before our parts are ready for synthesis and that is – double checking!
EPO with BbsI restriction sites and overhangs added:
AAA AGA AGA CAT AAT GGG CGT GCA CGA GTG CCC GGC GTG GCT GTG GCT GCT GCT GAG CCT GCT GAG CCT GCC GCT GGG CCT GCC GGT GCT GGG CGC GCC GCC GCG CCT GAT CTG CGA CAG CCG CGT GCT GGA GCG CTA CCT GCT GGA GGC GAA GGA GGC GGA GAA CAT CAC GAC GGG CTG CGC GGA GCA CTG CAG CCT GAA CGA GAA CAT CAC GGT GCC GGA CAC GAA GGT GAA CTT CTA CGC GTG GAA GCG CAT GGA GGT GGG CCA GCA GGC GGT GGA GGT GTG GCA GGG CCT GGC GCT GCT GAG CGA GGC GGT GCT GCG CGG CCA GGC GCT GCT GGT GAA CAG CAG CCA GCC GTG GGA GCC GCT GCA GCT GCA CGT GGA CAA GGC GGT GAG CGG CCT GCG CAG CCT GAC GAC GCT GCT GCG CGC GCT GGG CGC GCA GAA GGA GGC GAT CAG CCC GCC GGA CGC GGC GAG CGC GGC GCC GCT GCG CAC GAT CAC GGC GGA CAC GTT CCG CAA GCT GTT CCG CGT GTA CAG CAA CTT CCT GCG CGG CAA GCT GAA GCT GTA CAC GGG CGA GGC GTG CCG CAC GGG CGA CCG CGG TTC GNN GTC TTC AAA A
6. IN SILICO CLONING
Before sending any parts off to a DNA synthesis company, let’s first make sure that it will actually behave the way we want it to and that nothing went wrong during the design process. The best way to do this is to employ a software tool of choice and perform an in silico digest and ligation of the designed part. For our EPO-B3_B4, we’ll first try and clone it into the respective L0 plasmid backbone, pAGM1287. For more information on which plasmid backbones are needed for what cloning position, check out the parts table at the very top of this page. Secondly, we are going to try and assemble our EPO part into a L1 construct and check the reading frame.
A. CLONING L0-EPO-B3_B4
For this in silico digest, simply digest both your genetic part as well as the plasmid backbone with BbsI. If the design is right, you should see your part being assembled into the backbone in the correct
direction, taking the place of the LacZ-alpha gene. This exchange is very important for the actual experimental cloning, because, after E. coli transformation, the presence or absence of functional beta-galactosidase acts as a powerful
selection tool when screening for transformants with successfully cloned plasmids.

B. CLONING L1-sAP-EPO-Strep8His
Here we want to test whether our basic part can assemble at its designated place in a L1 construct and maintain the fusion protein‘s reading frame. As already elaborated in step Nr.4,
we’ll pick our L0-EPO-B3_B4 to be fused with an N-terminal sAP secretion tag and a C-terminal Strep8His - tag for purification. You can find all the sequences to our Moclo Mania parts in the iGEM Parts Registry. Click here for a comprehensive list of all our L0 parts available. Once all of them are within their respective L0 plasmid backbones, simply digest all the parts
you want to include in your construct plus our L1 destination vector named weird_plex with BsaI and let the MoClo magic happen. You should end up with what is depicted below, where once again, the
lacZ-alpha fragment of weird_plex has been replaced with your genetic construct. And ideally, your newly designed part should be placed comfortably in the cloning position that it was assigned. Last but not least – have your software tool
translate the fusion protein, beginning from the very first base triplet of the sAP secretion tag. Ideally, your translated sequence will stretch uninterruptedly across the entire length of your genetic construct only to come to a halt when encountering with the stop codon in B5.
Is this the case? Congratulations! You just created your first very own MoClo part! Now all that’s left to do is order it.

7. ORDER
Now that you have your readily designed and expertly tested MoClo part at hand, all there is left is to order it at your DNA synthesis company of choice! During our project, we relied on the synthesis services provided by IDT where small DNA pieces can be ordered as so-called gBlocks. If your genetic part reaches a certain size (which in the case of a protein like EPO, it definitely did), you might have to adjust your sequence towards the company’s standards in order to make it suitable for synthesis. This might include the elimination of repetitive sequences or GC-rich regions. IDT provides its very own software tool to alter your sequence accordingly, but most of the times it just comes down to tedious base per base typing work. Just be sure to always keep Leishmania’s codon usage table open in another tab and check back in with your amino acid sequence after you’re done. Depending on the size of your gene, it might even be necessary to split it up into separate subparts if you want to make use of a particular sequencing offer. In this case, make sure to equip these subparts with terminal BbsI recognition sites and adjust the separation region in order to allow for seamless reassembly - much like we did with our terminal sites in step Nr. 5! Be careful not to enter any BbsI or BsaI sites during this editing step! If you have any further questions or need help designing your part, please feel free to reach out to us! We are happy to help!
