
Theory
Phosphate pollution
 Phosphate levels in rivers and lakes
across Europe are highly variable. Phosphate concentrations
are at a normal level in mountainous areas of Scandinavia
or the Alps where there is no agricultural production. The
water bodies in fertile agricultural areas are however
heavily polluted as a result of extensive use of
fertilizers. Phosphates from fertilizers are largely
absorbed by the plants, but a significant proportion is
also drained into the rivers and lakes. There they help to
establish suitable conditions for the growth of undesirable
aquatic microorganisms such as cyanobacteria. [1] In Czech
Republic, higher phosphate concentrations in rivers and
lakes is also a hot topic. Based on the concentration of
phosphorus in the water, a waterbody is assigned a quality
class. Standard levels of phosphates correspond to classes
one and two, i.e. < 0.05 mg/ml and < 0.15 mg/ml
respectively. All rivers with average total phosphorus
concentrations above 0.15 mg/ml are considered
over-polluted. Almost half of all measured sites (441 out
of 919) were recorded to be over-polluted in 2012.
[2]
Phosphate levels in rivers and lakes
across Europe are highly variable. Phosphate concentrations
are at a normal level in mountainous areas of Scandinavia
or the Alps where there is no agricultural production. The
water bodies in fertile agricultural areas are however
heavily polluted as a result of extensive use of
fertilizers. Phosphates from fertilizers are largely
absorbed by the plants, but a significant proportion is
also drained into the rivers and lakes. There they help to
establish suitable conditions for the growth of undesirable
aquatic microorganisms such as cyanobacteria. [1] In Czech
Republic, higher phosphate concentrations in rivers and
lakes is also a hot topic. Based on the concentration of
phosphorus in the water, a waterbody is assigned a quality
class. Standard levels of phosphates correspond to classes
one and two, i.e. < 0.05 mg/ml and < 0.15 mg/ml
respectively. All rivers with average total phosphorus
concentrations above 0.15 mg/ml are considered
over-polluted. Almost half of all measured sites (441 out
of 919) were recorded to be over-polluted in 2012.
[2]
At some sites, the concentrations can go up to around 2.7 mg/ml, which is 18 times higher than the permissible limit. The annual average values of total phosphorus in the water of highly polluted streams can reach up to 0.8 mg/ml.
Wastewater treatment plants are addressing this problem, but not all are able to extract phosphorus from the water. Therefore, implanting an additional treatment device - phoscage - in rivers or lakes to accumulate exes phosphate could improve the situation. [3]
Bacterial Microcompartments
BMC are usually divided into two categories - ones which
are involved in anabolic and ones which are involved in
catabolic processes. The only currently known anabolic BMC
is carboxysome which is produced by cyanobacteria and some
chemotrophic bacteria. Carboxysomes enclose carbonic
anhydrase and RuBisCo and they fix carbon dioxide in the
Calvin-Benson-Bassham cycle. Catabolic BMCs or
metabolosomes are typically involved in metabolizing
different organic compounds. [8]
Bacterial microcompartments or BMC are large self-assembling polyhedral multi-protein structures. They typically consist of two main components - a selectively permeable protein shell and the enzymatic core. They are produced by a wide variety of bacteria and can carry a similar function to many eukaryotic organelles as they allow for certain biochemical reactions to be enclosed in an environment which is distinct and separated from bacteria's cytoplasm. These reactions can thus take place under very specific favourable conditions, the bacterial cell is protected from harmful metabolic intermediates and the intermediates can only partake in one biochemical reaction as they remain enclosed inside the BMC. [8]
There are three main types of shell proteins in BMCs - BMC-H, BMC-T and BMC-P. BMC-H are the most abundant and form cyclic hexamers. BMC-T form trimers. BMC-P is a minor component of the shell and they form the pentameric points of the polyhedral structure. BMC-P are also vital for the permeability of the shell. While there are many variations in BMCs across the bacterial kingdom, these basic structural components seem to be rather conserved. [8]
So how is this large structure put together? This process has been thoroughly described in carboxysomes but still needs to be explored further in catabolic BMCs. In α-carboxysomes, the enzymatic core and the shell assemble simultaneously whereas in β-carboxysomes, the enzymatic core is assembled first and is then enclosed by the shell. As the core enzymes of many metabolosomes tend to aggregate, it is assumed that metabolosomes likely assemble in a similar fashion to β-carboxysomes. Many enzymes from the core contain an extension, which interacts with the inside parts of the shell. [8]
BMC type which is particularly important for our project is the catabolic propanediol utilization BMC (Pdu). Pdu operon was recently found in P. thermoglucosidasius which is quite closely related to our bacteria of choice - B. subtilis. This operon includes 17 genes coding various proteins from the shell as well as the enzymatic core. In a recent study, five essential shell proteins have been identified. By connecting the extension of one of the enzymatic components of the Pdu operon to a different enzyme, it should be possible to enclose this enzyme inside of the BMC instead. [7,9]
The Pho pathway
Bacteria metabolize phosphorus in the form of the orthophosphate anion (PO43-), also known as inorganic phosphate (Pi). Since Pi plays a role in a variety of processes which are vital for any living organism, every living thing must have developed some physiological ability to acquire and store Pi in order to survive in a Pi-deficient environment. This ability in most bacteria is regulated by the phosphate (Pho) pathway. Pho pathway is activated when Pi levels are low and it signals for the expression of several specific operons involved in saving up and efficiently utilizing Pi. [10]
Regulation of the Pho pathway has been first characterized in Escherichiacoli. In E. coli, this pathway consists of a two-component PhoB/PhoR system. PhoR is a sensor histidine-kinase protein anchored to the inner membrane. PhoB regulates the transcriptional response inside of the cell. In low Pi concentrations, PhoB is phosphorylated by the sensor kinase PhoR. This allows the phosphorylated response regulator to interact with promoters (such as PPho) and initiate gene transcription. In addition, the Pi-sensing pathway includes the Pst transporter (PstSCAB) and the metal binding protein PhoU which dephosphorylates PhoB in a Pi-rich environment. [10,11]
The Pho pathway has currently also been characterized in
other bacteria. In Bacillus subtilis, Pi management
follows the same principles as in E. coli, but
differs in the structure of the Pho pathway itself. B.
subtilis does not encode a homologue of the
metal-binding PhoU protein and the main regulator of the
transcription of genes under the Pho promotor is the
two-component protein system PhoP/PhoR. [12]
The most common biological components that are activated by the Pho pathway include extracellular enzymes capable of extracting phosphate from organic compounds, specific transporters or enzymes involved in Pi storage. The promoters of these genes have rather similar sequences and some sites are even fully conserved. [10,13]
In our project PHOSCAGE, we decided to use this pathway activated by low phosphate concentrations to ensure that BMC production only takes place at a threshold phosphate levels and thus to relieve the burden on bacterial metabolism. For this purpose, we used the promoter of the phoA gene (alkaline phosphatase A) from B. subtilis and placed a repressor which blocks the promoter of our BMC genes under its control. The phoA gene promoter was chosen as it has a known sequence and by comparing transcriptional values of Pho regulon-induced genes in the http://subtiwiki.uni-goettingen.de/ database. [14]
The phosphate metabolism of Bacillus subtilis differs quite a lot from the more familiar phosphorus metabolism of other organisms. Bacillus subtilis and other gram positive bacteria do not produce polyphosphate kinase 1 (PPK1) or polyphosphate kinase 2 (PPK2) which catalyse the formation of long polyphosphate chains. Thus, a different phosphate accumulation system is present. Bacillus subtilis stores phosphates by forming a phosphate-rich glycopolymer - wall teichoic acid (WTA) - which is incorporated in their cell wall. This biochemical reaction is mediated by the TagF protein, and appears to use CTP as a source of phosphates rather than ATP which is utilised by PPKs. In the absence of phosphates, WTA is digested by phosphodiesterases GlpQ and PhoD and phosphates are released into cytoplasm. The decomposition of WTA is controlled by the PhoPR complex and WTA is then replaced by teichuronic acid which does not contain phosphates. This should not negatively affect the lifespan of Bacillus subtilis. [16,18]
In our project, we would like to enclose suitable PPK in the Pdu BMC so that the resulting polyphosphate chain is protected from phosphodiesterases and the rate of phosphate accumulation is increased. This however begs the question of which criteria to look at when selecting a suitable PPK. First and foremost is the rate of polyphosphate production by PPK, which would make the most suitable PPK from Corynebacterium glutamicum or from Escherichia coli. However, upon closer examination, most PPKs with higher activity work as multi-mers, where some of the subunits might also have different functions. This could be a major problem when attempting to encapsulate this kinase into BMCs, where misfolding could occur. For this reason, we searched for a monomeric PPK derived from Propionibacterium shermanii. It should however be noted that successful encapsulation of the multimeric PPK from Escherichia coli into BMCs was already described in the Liang 2017 study so it might be an issue. [9,17,19]
Biofilm
Biofilm is a structure formed by a larger community of bacteria, either consisting of a single species or of several species. Biofilm is formed by the settling (adhesion) of planktonic bacteria on a surface. These bacteria proliferate and secrete a variety of substances such as proteins, glycoproteins and polysaccharides to form an extracellular envelope. The growth of a biofilm can be imagined as a small settlement evolving and changing into a large city. In our hypothetical settlement, there are bacteria that more or less all have the same task - namely to multiply and produce useful substances - which is quite demanding and sometimes leaves them with not enough energy left to defend themselves against external influences. In a large city, a process akin to differentiation is already taking place. In other words, even bacteria of the same species perform different tasks based on their position in biofilm. Surface bacteria maintain the stability of the structure and form tunnels for nutrient uptake. Inner bacteria focus more on nutrient metabolism so that they can provide energy to other bacteria that serve other functions like antibiotic resistance. Many others then focus on increasing resistance to biological agents (like antibiotics). [21]
 As the biofilm matures, it begins to
release planktonic bacteria outside at a higher rate, which
can lead to the establishment of new settlements
(colonies). Communication within biofilm is mediated by the
exchange of chemical signals, the best known being quorum
sensing (QS). The concentration of QS signaling molecules
informs the bacteria about the population density in the
biofilm. [22]
As the biofilm matures, it begins to
release planktonic bacteria outside at a higher rate, which
can lead to the establishment of new settlements
(colonies). Communication within biofilm is mediated by the
exchange of chemical signals, the best known being quorum
sensing (QS). The concentration of QS signaling molecules
informs the bacteria about the population density in the
biofilm. [22]
Living in a biofilm brings the possibility of division of labour and thus more energy can be allocated to each individual process while benefiting the entire population. Biofilm structure is also much more resistant to mechanical damage and has increased ability to resist antibiotics. Another benefit of biofilms is greater efficiency of horizontal gene transfer which can be up to 1000 times better than in the planktonic state [21]. It is also quite interesting that the bacterium retains these properties for a short time after leaving the biofilm.
In our project, we are considering building a consortium of native bacilli which are able to produce biofilm and our modified B. subtilis which are not and can benefit from being part of the biofilm without having to spend energy maintaining it. We expect this to improve the vitality of our modified bacteria, leaving them with more energy to produce our BMCs.
Bacillus subtilis
Bacillus subtilis is a very commonly used model organism for Gram-positive bacteria. Its rod-shaped cells are usually 4-10 μm long and their diameter varies between 0,25 and 1 μm. It is able to form endospores in adverse environments and thus is able to persevere until conditions improve. It is usually found in soil and in the gastrointestinal tract of some mammals, including humans. Bacillus subtilis is widely used in molecular biology and biotechnology and it is considered to be somewhat of a Gram-positive equivalent of Escherichia coli.
We chose Bacillus subtilis because of its GRAS status as well as its ability to thrive in lower temperatures. The number and variety of genetic tools available was also a point in its favour. Additionally, we were already familiar with this organism from last year's igem project and we also had some material left from our experiments.
 Last year we approached Dr. Libor
Krásný and his colleagues from the Laboratory of Microbial
Genetics and Gene Expression which operates under the Czech
Academy of Science in Prague. He gave us a lot of helpful
tips and instructions to help us work with this bacterium
and he also provided us with B. subtilis cells which
we have used in our project. We consulted parts of our
project with Dr. Krásný this year as well. We were very
grateful for his help.
Last year we approached Dr. Libor
Krásný and his colleagues from the Laboratory of Microbial
Genetics and Gene Expression which operates under the Czech
Academy of Science in Prague. He gave us a lot of helpful
tips and instructions to help us work with this bacterium
and he also provided us with B. subtilis cells which
we have used in our project. We consulted parts of our
project with Dr. Krásný this year as well. We were very
grateful for his help.
Vectors
We have consulted our project with the Laboratory of Medical Genetics and Gene Expression led by Dr. Krásný from Prague, who recommended using two plasmids as our synthetic vectors. These plasmids are specifically designed for ectopic integration of genes into the genome of Bacillus subtilis. We used vectors pDG1664 and pDG3661. Our chosen plasmids are so-called shuttle vectors, which means that they can be used in Escherichia coli as well as in Bacillus subtilis. Each of these two bacteria strains does however act a little differently therefore a different system for selection of transformed cells had to be used. The origin of replication is recognized only in E. coli and thus the plasmids cannot be replicated in B. subtilis . Therefore, these plasmids can be propagated as epistomes in E. coli, but fast and immediate integration into the B. subtilis chromosome must follow after transformation to ensure the genes for antibiotic resistance and our constructs are present. Double recombination events occur during the integration of these vectors.
In our experiments we used Escherichia coli strain DH5α commercial competent, E. coli competent cells provided by PROMEGA, E. coli expression cells BL21 (DE3) and Bacillus subtilis 168.
Every method mentioned here is also described in the Experiment section in greater detail.
pDG1664
 We used the same vectors as last
year as we have already had experience working with them.
One of them being the pDG1664 vector. This vector was used
to integrate one of our four constructs. The rest were
integrated using the pDG3661 vector mentioned
below.
We used the same vectors as last
year as we have already had experience working with them.
One of them being the pDG1664 vector. This vector was used
to integrate one of our four constructs. The rest were
integrated using the pDG3661 vector mentioned
below.
This plasmid contains three genes for antibiotic resistance - gene erm (resistance to erythromycin), gene spc (resistance to spectinomycin) and gene bla (resistance to ampicillin). The gene of interest is cloned into restriction sites for BamHI and HindIII located inside of the thrC gene. This gene is responsible for chromosomal integration in Bacillus subtilis. Transformants of Escherichia coli can be detected by their resistance to ampicillin. Successfully transformed cells of Bacillus subtilis were selected using a combination of antibiotics erythromycin and lincomycin. This is known as MLS selection. We then used PCR and sequencing to prove the successful integration of the pDG1664 vector with our construct. [23, 24]
pDG3661
Vector pDG3661 is 10 407 bp long and it was used for the
integration of constructs C and D. This plasmid also
contains the spoVG-lacZ gene, which was created by the
fusion of genes spoVG and lacZ from Bacillus
subtilis. This sequence can be replaced by different
recombinant DNA using the BamHI restriction site. The
selection of transformed Escherichia coli is done
using the bla gene which codes ampicillin resistance. Spc
gene and catgene, which encode resistance to antibiotic
chloramphenicol, are used for the selection of
transformed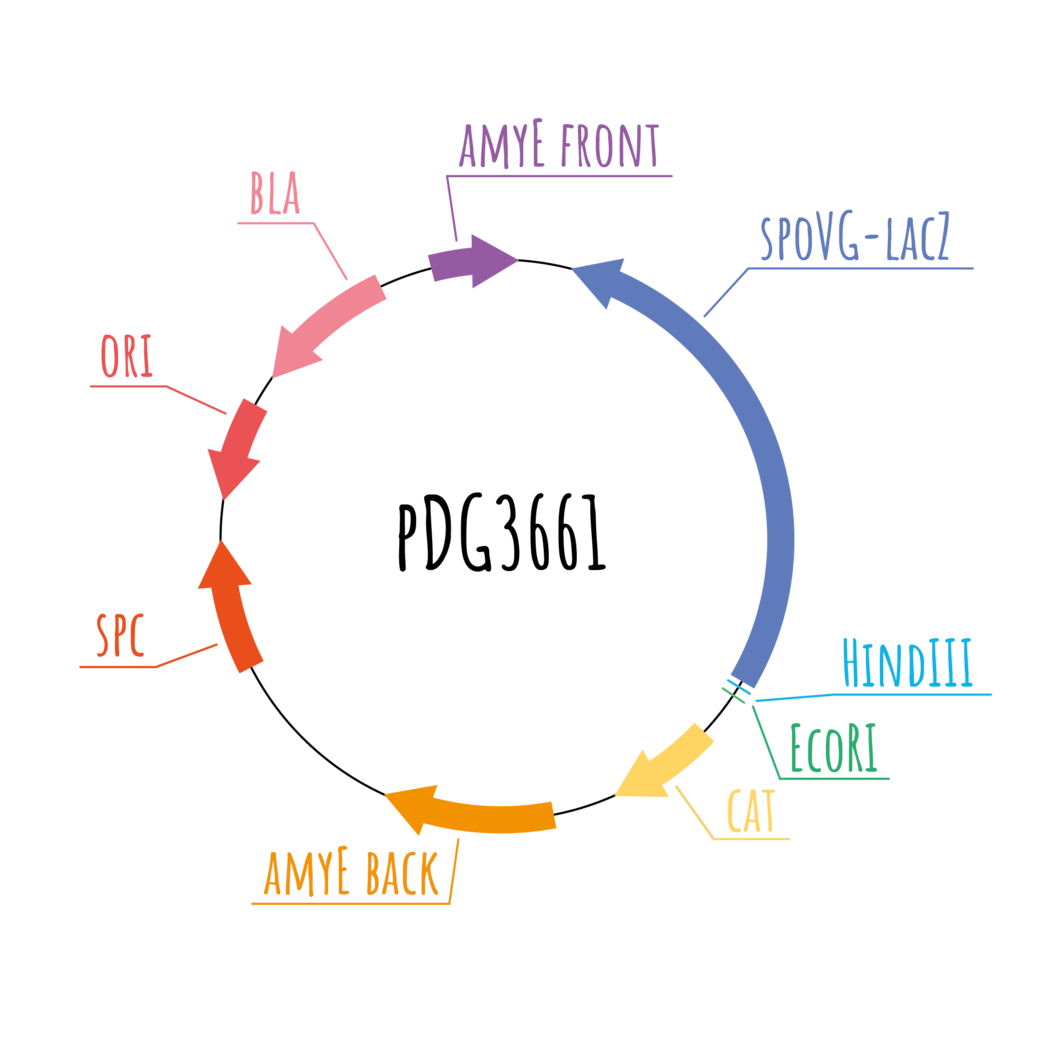 Bacillus subtilis cells. The
integration of this vector takes place in the amyE sequence
so the cat gene and 3060 bp long spoVG-lacZ gene are
inserted with the synthetic constructs C and D.
Bacillus subtilis cells. The
integration of this vector takes place in the amyE sequence
so the cat gene and 3060 bp long spoVG-lacZ gene are
inserted with the synthetic constructs C and D.
We used restriction digestion to verify the cloning of our insert into this plasmid. More information about this method can be found in the section experiments .
We also used the AmyE test to demonstrate the integration of the vector with corresponding construct into the Bacillus subtilis chromosome. Constructs C and D were inserted into the sequence of AmyE gene, which encodes α-amylase. When the AmyE sequence is left intact, the starch in agar medium is gradually digested by this enzyme. The addition of a few iodine crystals to the surrounding medium makes this process visible in the form of a clear halo surrounding the colonies. In the case of a successful integration into AmyE, the bacteria cannot produce α-amylase and therefore this halo effect does not occur. More about using restriction digestion and AmyE test can be found in the section experiments . [25,26]
IDT plasmids
We ordered our 4 synthetic constructs from the company IDT, through the iGEM sponsorship. We opted to have our synthetic constructs, A, B, C and D incorporated in commercial pUCIDT plasmids.
This plasmid contains the gene for resistance to the antibiotic ampicillin and has a replication origin for replication in E.coli.
Sources:
-
European Environment Agency. 2019. Nutrients in Freshwater in Europe. https://www.eea.europa.eu/data-and-maps/indicators/nutrients-in-freshwater/nutrients-in-freshwater-assessment-published-10
-
Water quality assessment. 2015. Czech Hydrometeorological Institute. https://voda.gov.cz/portal/isvs/chmu/jvp/cz/
-
Classification of surface water quality. Water Management Research Institute. https://www.vtei.cz/2017/12/klasifikace-kvality-povrchovych-vod/
-
Reinhard, C., Planavsky, N., Gill, B. et al. Evolution of the global phosphorus cycle. Nature. 541, 386–389 (2017). https://doi.org/10.1038/nature20772
-
Phosphorus: Essential to Life—Are We Running Out?. State of the Planet [online]. Available at: https://news.climate.columbia.edu/2013/04/01/phosphorus-essential-to-life-are-we-running-out/
-
Yuan, Z., Jiang, S., Sheng, H., Liu, Xin, Hua, H., Liu, Xuewei, Zhang, Y., 2018. Human Perturbation of the Global Phosphorus Cycle: Changes and Consequences. Environ. Sci. Technol. 52, 2438–2450. https://doi.org/10.1021/acs.est.7b03910
-
Wade Y., Daniel R. A., Leak D. J. 2019. Heterologous Microcompartment Assembly in Bacillaceae: Establishing the Components Necessary for Scaffold Formation. ACS Synth. Biol. 8: 1642-1654
-
Karfeld C. A., Aussignargues C., Zarzycki J., Cai F., Sutter M. 2018. Bacterial microcompartments. Nat. Rev. Microbiol. 16 (5): 277-290. nbspdoi:10.1038/nrmicro.2018.10.
-
Liang M., Frank S., Lünsdorf H., Warren M. J., Prentice M. B. 2017. Bacterial microcompartment-directed polyphosphate kinase promotes stable polyphosphate accumulation in E. coli. Biotechnol. J. 12: 1600415. DOI 10.1002/biot.201600415.
-
Santos-Beneit F. 2015. The Pho regulon: a huge regulatory network in bacteria. Front Microbiol. 6: 402. doi: 10.3389/fmicb.2015.00402
-
Wanner B. L., & Chang B. D. 1987. The phoBR operon in Escherichia coli K-12. J Bacteriol. 169(12): 5569–5574. doi: 10.1128/jb.169.12.5569-5574.1987
-
Vershinina O. A., & Znamenskaya, L. V. 2002. The Pho Regulons of Bacteria. Microbiology. 71: 497–511. doi: 10.1023/A:1020547616096
-
Eder S., Shi L., Jensen K., Yamane K., Hulett F. M. 1996. A Bacillus subtilis secreted phosphodiesterase/alkaline phosphatase is the product of a Pho regulon gene, phoD. Microbiology (Reading). 142 ( Pt 8): 2041-7. doi: 10.1099/13500872-142-8-2041
-
http://subtiwiki.uni-goettingen.de/
-
Balasubramanian S., Aubin-Tam M.-E., Meyer A. S. 2019. 3D Printing for the Fabrication of Biofilm-Based Functional Living Materials. ACS Synth. Biol. 8: 1564-1567. 10.1021/acssynbio.9b00192
-
DEVINE, Kevin M. Activation of the PhoPR-Mediated Response to Phosphate Limitation Is Regulated by Wall Teichoic Acid Metabolism in Bacillus subtilis. Frontiers in Microbiology [online]. 2018, 9 [cit. 2021-10-12]. ISSN 1664-302X. doi:10.3389/fmicb.2018.02678
-
ZHANG, H., K. ISHIGE a A. KORNBERG. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proceedings of the National Academy of Sciences [online]. 2002, 99(26), 16678-16683 [cit. 2021-10-12]. ISSN 0027-8424. doi:10.1073/pnas.262655199
-
Pepin CA, Wood HG. The mechanism of utilization of polyphosphate by polyphosphate glucokinase from Propionibacterium shermanii. J Biol Chem. 1987 Apr 15;262(11):5223-6. PMID: 3031045.
-
PEREIRA, Mark P., Jefferey W. SCHERTZER, Michael A. D'ELIA, Kalinka P. KOTEVA, Donald W. HUGHES, Gerard D. WRIGHT a Eric D. BROWN. The Wall Teichoic Acid Polymerase TagF Efficiently Synthesizes Poly(glycerol phosphate) on the TagB Product Lipid III. ChemBioChem [online]. 2008, 9(9), 1385-1390 [cit. 2021-10-12]. ISSN 14394227. doi:10.1002/cbic.200800026
-
LINDNER, Steffen N., Dolores VIDAURRE, Sabine WILLBOLD, Siegfried M. SCHOBERTH a Volker F. WENDISCH. NCgl2620 Encodes a Class II Polyphosphate Kinase in Corynebacterium glutamicum. Applied and Environmental Microbiology [online]. 2007, 73(15), 5026-5033 [cit. 2021-10-12]. ISSN 0099-2240. doi:10.1128/AEM.00600-07
-
SOLANO, Cristina, Maite ECHEVERZ a Iñigo LASA. Biofilm dispersion and quorum sensing. Current Opinion in Microbiology [online]. 2014, 18, 96-104 [cit. 2021-10-20]. ISSN 13695274. doi:10.1016/j.mib.2014.02.008
-
COENYE, T. Biofilms. Brenner's Encyclopedia of Genetics [online]. Elsevier, 2013, 2013, s. 335-337 [cit. 2021-10-20]. ISBN 9780080961569. doi:10.1016/B978-0-12-374984-0.00154-6
-
Guérout-Fleury A. M., Frandsen, N., and Stragier, P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180(1-2): 57–61. DOI: 10.1016/s0378-1119(96)00404-0
-
https://www.ncbi.nlm.nih.gov/nuccore/1185593
-
Krásný, L., and Gourse, R. L. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. The EMBO journal 23(22): 4473–4483. DOI: 10.1038/sj.emboj.7600423
Igem Team Brno, Czech Republic 2021
