PROJECT
Design
INTRODUCTION
A critical barrier in predicting patient response to immunotherapy is the lack of reliable predictive biomarkers [1, 2] One factor that impacts immunotherapy outcome is the immune activity in the tumour microenvironment [2]. To support the discovery and validation of predictive biomarkers, we set out to design an in-vivo biomarker profiling tool to characterize tumour microenvironments in mouse models.

Few signaling pathways in bacteria are known to respond to human biomarkers. To ensure that our tool would be able to detect relevant biomarkers in the TME, we designed a functional screening platform to find promoters responsive to our biomarkers of interest. This is our promoter discovery pillar.
Through discussions with cancer researchers and oncologists, we confirmed that there is an unmet need to better characterize tumour microenvironments and their heterogeneity across individuals. We developed a biosensor to profile tumours for in-vivo implementation through generating a reporter upon immune activity in the tumour. Better characterization of tumour upon treatment will aid in the search for predictive biomarkers for therapy outcome, and it would aid in search for predictive biomarkers. This is our tumour microenvironment (TME) profiling pillar.
Finally, to ensure a relatively convenient diagnostic platform, we designed a workflow to detect immune biomarkers in-vivo and release reporters that can be recovered in urine. This approach would reduce costs and suffering relative to other approaches, and this is our urine detection pillar.
CHASSIS: SALMONELLA
During our initial ideation process, we found that bacteria like Salmonella typhimurium can specifically colonize tumour sites and not healthy tissues [3-5]. Among these tumour-colonizing bacteria, Salmonella is the most widely studied due to ease of genetic manipulation [1]. The hypoxic and lactate-rich environments in tumours act as chemotactic factors to attract Salmonella to tumour sites, whereas normal tissues do not support Salmonella growth [1]. Since the discovery of tumour-targeting Salmonella in 1997, there has been extensive research into using Salmonella as a novel cancer therapy [1, 4]. It has been shown that Salmonella can persist at tumour sites for 4-8 weeks, and there have been clinical trials for melanoma, renal cell carcinoma, liver cancer, and so on where Salmonella was applied as a therapy [5-8].
We spoke to Dr. Saltzman, a clinician-researcher in Salmonella therapy, to learn about how Salmonella are used in clinical trials. He recommended that we look into minimally immunogenic strains of Salmonella. More immunogenicity would provoke unwanted immune responses in other areas of the body which would lead to bad side effects. Salmonella-induced toxicity in humans and mice is also affected by the bacteria’s route of administration. Intravenous injection results in higher tumour colonization while higher toxicity; whereas oral administration has lower toxicity and lower colonization. Learning this from Dr. Saltzman helped us decide that intravenous administration may suit our need as a start since higher colonization efficiency could facilitate characterizing the biosensor’s function. (Implementation).
The tumour-targeting and colonizing features of Salmonella, its ease of genetic manipulation, and its use in previous clinical trials make it a suitable chassis for our purpose.
To seek a suitable strain of Salmonella and learn more about safety practices, we reached out to world-experts on Salmonella at Finlay laboratory at UBC. They provided us with a highly attenuated strain called SL1344 ∆invA∆ssaR double mutant. Key components of the Salmonella pathogenicity island-1 (invA gene) and SPI-2 type III secretion system (ssaR gene) used by Salmonella to evade host responses and induce pathogenicity are deleted in this strain through allelic exchange and a suicide vector [10], reducing pathogenicity of Salmonella. For our safety practice of using Salmonella, please see our Safety page.
SUBSTRATE-INDUCED GENE EXPRESSION (SIGEX) FOR DISCOVERY OF NOVEL PROMOTERS
Different cancer types may have different immune markers of interest, and as research advances, more biomarkers will surely be discovered. Also, many human biomarkers aren't recognized by any known bacterial signalling systems. For these reasons, we aimed to make our system adaptable to other inducible promoters by screening across the Salmonella genome for regulatory elements that are responsive to biomarkers of interest.
First, Salmonella's ability to target tumours rely on their ability to detect and respond to chemotactic factors in the TME. Secondly, previous efforts by Leschner et al (2012) and Arrach et al (2008) have demonstrated that the Salmonella genome harbors promoters that are inducable by the tumour environment. In particular, Leschner ’s study shows that there are hypoxia-responsive promoters in the Salmonella genome.
There is limited work on further exploring the roles of specific TME biomarkers in inducing gene expression in Salmonella. Our promoter discovery component will address this knowledge gap.
1) To construct a screening library of sheared Salmonella genome fragments
2) To construct a screening vector that could discern regulatory elements with directionality
3) To perform proof-of-concept screening of a substrate of interest
Construct Development
We implemented a promoter trap adapted from the design from UBC iGEM 2019 and Bumann and Valdivia (2007) [8]. In a promoter-less vector, we inserted a restriction site (BamHI) upstream of a forward reporter (GFP) and a reverse reporter (RFP). The restriction site is where fragments from the sheared Salmonella genome will be inserted. If this inducible promoter happens to have directionality and regulates in the forward direction, it will drive the expression of GFP, and if it regulates the reverse direction, it will drive the expression of the reverse RFP. These results can be sorted by fluorescence colour, resulting in candidate inducible promoters that can be further validated through sequencing.
The extracted Salmonella genome is sheared to 500-700bp with a sonicator as specified in Bumann and Valdivia’s (2007) method.

BIOMARKER SELECTION
We spoke to cancer immunologists and oncologists (Integrated Human Practices) to learn about the biomarkers that would be most useful to detect for predicting immunotherapy outcomes.
According to Dr. Morgan Roberts, Dr. James Lim, and others, biomarkers such as high tumour mutation burden (TMB), high expression of tumour surface protein PD-1, or T cell surface protein PD-L1, are relatively characteristic of immune-active tumours but are not strongly predictive of immunotherapy response in many cases. They suggested against relying on only one biomarker to predict treatment outcome. Therefore, we set out to design a genetic AND-gate as a starting point for multiplexing multiple biomarkers in our biosensor.
There are many biomarkers within the TME that are relevant for immune activities. Our proof-of-concept focuses on TNFa and lactate, because there are known regulatory bacterial elements that can respond to these biomarkers, and both are clinically informative on TME immune activity as well as hypoxia levels.
TNFa
In our research, we found that Tumour Necrotic Factor a (TNFa) plays an essential role in diverse cellular events involving immune cells within the TME (Laha et al, 2021). We also found that TNFa can upregulate production of Salmonella’s invasion protein SipA (Ma et al, 2010; Laha et al, 2021). As such, we hypothesized there was a genetic regulatory pathway in Salmonella that was responsive to TNFa. Taken together, we decided to choose it as one of our proof-of-concept biomarkers.
Lactate
We chose lactate as the other biomarker-of-interest. Lactate is enriched in the tumour environment as a byproduct of anaerobic respiration, and it negatively regulates immune cell functioning in tumours (Wang et al, 2020). It not only impairs monocyte differentiation into dendritic cells (DCs) and further decreases their antigen-presentation functions, lactic acid facilitates the infiltration of immunosuppressive cell types and contributes to cancer immune escape (Wang et al, 2020). Considering there is a known lactate-inducible regulatory mechanism within Salmonella, we decided to focus on lactate as the other biomarker.
Because Salmonella preferentially colonizes tumours and not healthy tissues, the signals we receive would theoretically only be released by tumour-related activities [4]. If control experiments in mice show that there is a background signal from healthy tissues, this could be controlled for substracting the background noise from the obtained biosensor output.
A GENETIC AND-GATE TOOL
In seeking a suitable reporter to use in the tumour environment, we wanted the readout to be easily detectable using simple devices, and for the signal to noise ratio to be low meaning that a large amount of reporter needs to be produced. There should also be low baseline activity, meaning a system foreign to Salmonella should be used so that the reporter signal is not confounded by normal Salmonella processes. Another design consideration was that the cost of running the screen should be low, and ideally we would require only the substrate of interest and no other input for signal generation.
In our research, we found that bacterial lux operons are used for in-vivo localization of bacteria in mouse models and the bioluminescence signal can be detected through bioluminescence imaging devices [15]. The bacterial lux operon consists of six proteins, luxA-G. luxA and luxB form a heterocomplex and function as a luciferase enzyme; luxC, D, and E form a fatty acid reductase [16]. There is no additional input needed for producing the luciferin reporter [16]. Therefore, we decided a readout of bioluminescence would suit our needs given its ease of detection and high signal to noise ratio.
1) To characterize the function of the TNFa-inducible promoter sicA and lactate-inducible promoter plldR with dose response experiments at physiological concentrations
2) To create E. coli strains that express half-operons under the control of inducible promoters
3) To create Salmonella strains that express half-operons under the control of inducible promoters
4) To double transform Salmonella with the two half-operons and validate its function as an AND-gate
Construct Development
To design the genetic AND-gate, we wanted to split the operon such that luxA, luxB, and luxG are expressed when one biomarker is present, and the other half is expressed when the other is present, such that light is produced only when both biomarkers are present.
To allow easy adaptation to different biomarkers, the inducible promoters are specific to the biomarker of interest. In our proof-of-concept construction, we picked plldR, a lactate inducible unit, and psicA, the promoter of the sipA gene which is hypothesized to be regulated by TNFa.
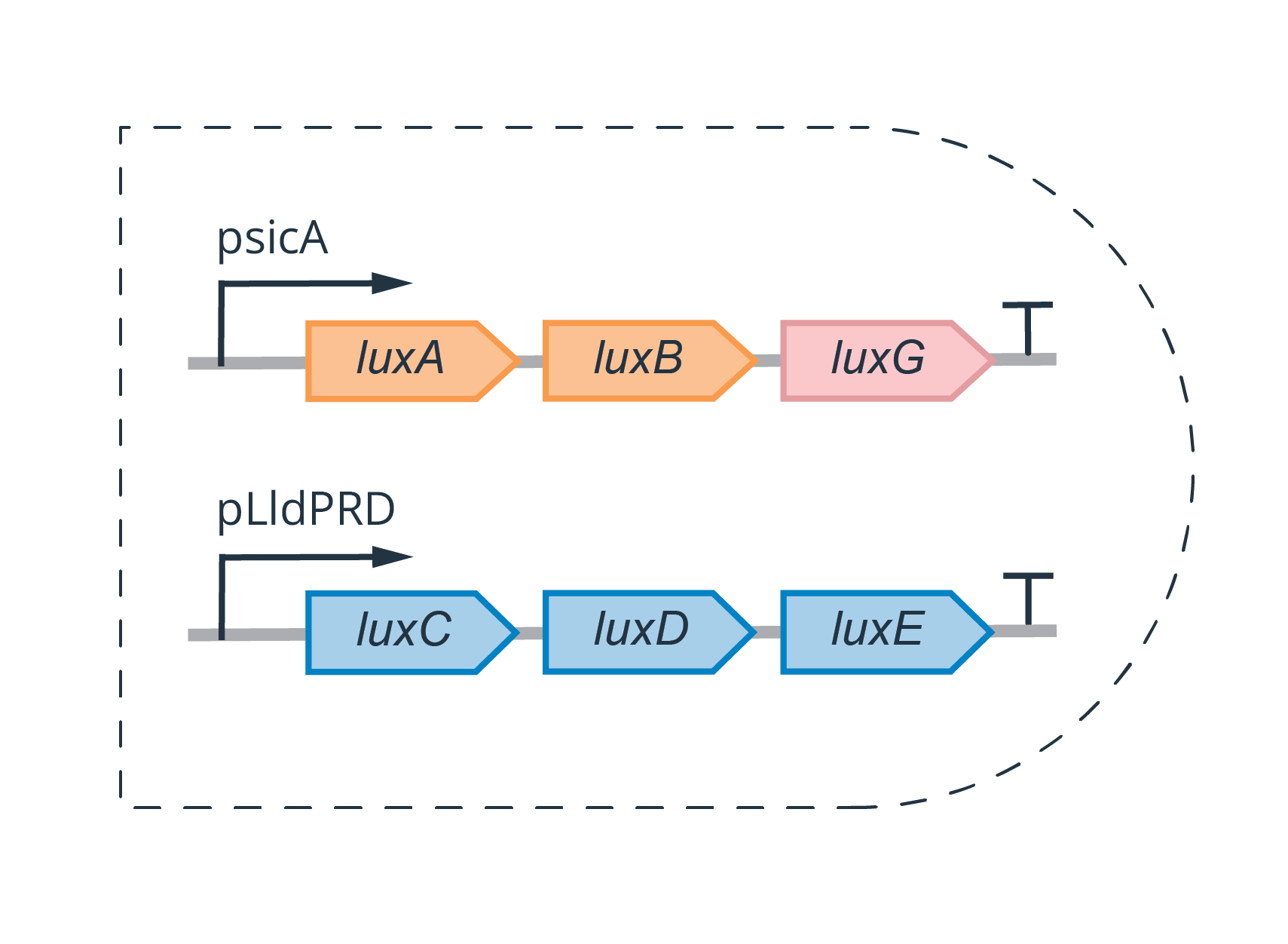
For details on experiment planning and execution, please refer to our Experimental Design page.
URINE DETECTION
While bioluminescence from the lux operon is ideal as a characterization readout in mice models, bioluminescent imaging is not yet possible for human tumours. In order to make the system clinically applicable, we need to use another reporting system that can be recovered in urine. Unfortunately, detection of bacterial luciferin in urine has not yet been demonstrated.
However, luciferin detection in urine is possible. We found research from Danino et al, that created an E.coli biosensor for urine detection of cancer. In their work, urine detection was achieved through additional administration of LuGal, a conjugate of galactose and luciferin, and an engineered E.coli expressing beta-galactosidase [4].
Inspired by Danino’s work [4], we designed a similar promoter-based AND-gate system for urine signal detection, but using a split protein and a protein-protein interaction technique.

For the split protein, we will split beta-galactosidase protein into an α and an ω fragment. Indonesia iGEM 2013 (biobricks) has executed and demonstrated the functionality of the α and ω fragment biobricks.
To ensure that the two halves of the protein interact to form a functional beta-galactosidase, we draw inspiration from a natural protein-protein interaction between SipA, an invasion protein in Salmonella, and InvB, a type III secretion-associated chaperone [9, 17]. InvB interacts with SipA to assist the secretion of SipA [17]. Their interaction has been characterized via a split luciferase [17].
For our purpose, we will leverage both SipA-InvB interactions together with a split beta-galactosidase. We will connect sipA with the α fragment of beta-galactosidase via a linker DNA and under the control of the first inducible promoter, and similarly, invB is linked to the ω fragment of beta-galactosidase, under the control of a second inducible promoter. This way, only the environment with both biomarkers will have LacZ production to release luciferin from the addition of LuGal. This signal has been shown to be recoverable through urine [4].
MICROENVIRONMENT AUGMENTING FUNCTIONALITY
As an additional functionality, we wanted to create genetically engineered Salmonella which could express and release molecules to modify the tumour microenvironment such that the tumour becomes more vulnerable to immunotherapy. For this aspect, we are collaborating with the iGEM HU Berlin team. Please refer to our Partnership page, and HU Berlin's wiki (Target Taxi) to learn more about the therapeutic potentials.
MODELLING THE TUMOUR MICROENVIRONMENT
In the current project, we are not able to access tumour organoids or mouse models to recapitulate a cancer microenvironment. To mimic the microenvironments that our biosensor will function in, we created a computational simulation of a tumour spheroid and its surrounding blood vessel network, building off of the work from Welter and Rieger (2016), Fredrich et al (2019) [18, 19]. To learn more about the software simulations, please refer to the Modelling page.

Despite our limited access to resources that would allow us to test our design in-vivo, we consulted with various clinical experts about the possible implementation of our system to the public. The climb to this final goal would involve rounds of in-vivo mouse and human testing, which is extensively documented on our Implementation page.
[1] McKean, W. B., Moser, J. C., Rimm, D., & Hu-Lieskovan, S. (2020). Biomarkers in precision cancer immunotherapy: promise and challenges. American Society of Clinical Oncology Educational Book, 40, e275-e291.
[2] Lei, Y., Li, X., Huang, Q., Zheng, X., & Liu, M. (2021). Progress and Challenges of Predictive Biomarkers for Immune Checkpoint Blockade. Frontiers in Oncology, 11, 609.
[3] Kwong, G. A., Ghosh, S., Gamboa, L., Patriotis, C., Srivastava, S., & Bhatia, S. N. (2021). Synthetic biomarkers: a twenty-first century path to early cancer detection. Nature Reviews Cancer, 1-14.
[4] Chien, T., Doshi, A., & Danino, T. (2017). Advances in bacterial cancer therapies using synthetic biology. Current opinion in systems biology, 5, 1-8
[5] Toso, J. F., Gill, V. J., Hwu, P., Marincola, F. M., Restifo, N. P., Schwartzentruber, D. J., ... & Rosenberg, S. A. (2002). Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology, 20(1), 142.
[6] Heimann, D. M., & Rosenberg, S. A. (2003). Continuous intravenous administration of live genetically modified salmonella typhimurium in patients with metastatic melanoma. Journal of immunotherapy, 26(2), 179-180.
[7] Nemunaitis, J., Cunningham, C., Senzer, N., Kuhn, J., Cramm, J., Litz, C., ... & Sznol, M. (2003). Pilot trial of genetically modified, attenuated Salmonella expressing the E. coli cytosine deaminase gene in refractory cancer patients. Cancer gene therapy, 10(10), 737-744.
[8] Gniadek, T. J., Augustin, L., Schottel, J., Leonard, A., Saltzman, D., Greeno, E., & Batist, G. (2020). A Phase I, Dose Escalation, Single Dose Trial of Oral Attenuated Salmonella typhimurium Containing Human IL-2 in Patients With Metastatic Gastrointestinal Cancers. Journal of Immunotherapy (Hagerstown, Md.: 1997), 43(7), 217.
[9] Danino, T., Prindle, A., Kwong, G. A., Skalak, M., Li, H., Allen, K., ... & Bhatia, S. N. (2015). Programmable probiotics for detection of cancer in urine. Science translational medicine, 7(289), 289ra84-289ra84.
[10] Caveney, N. A., Serapio-Palacios, A., Woodward, S. E., Bozorgmehr, T., Caballero, G., Vuckovic, M., Deng, W., Finlay, B. B., & Strynadka, N. C. (2020). Structural and cellular insights into the L,D-transpeptidase YcbB as a therapeutic target in Citrobacter Rodentium, salmonella typhimurium, and salmonella typhi infections. Antimicrobial Agents and Chemotherapy, 65(2).
[11] Pawelek, J. M., Low, K. B., & Bermudes, D. (1997). Tumor-targeted Salmonella as a novel anticancer vector. Cancer research, 57(20), 4537-4544
[12] Leschner, S., Deyneko, I. V., Lienenklaus, S., Wolf, K., Bloecker, H., Bumann, D., ... & Weiss, S. (2012). Identification of tumor-specific Salmonella Typhimurium promoters and their regulatory logic. Nucleic acids research, 40(7), 2984-2994.
[13] Arrach, N., Zhao, M., Porwollik, S., Hoffman, R. M., & McClelland, M. (2008). Salmonella promoters preferentially activated inside tumors. Cancer research, 68(12), 4827-4832.
[14] Clairmont, C., Lee, K. C., Pike, J., Ittensohn, M., Low, K. B., Pawelek, J., ... & Zheng, L. M. (2000). Biodistribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of Salmonella typhimuvium. Journal of Infectious Diseases, 181(6), 1996-2002.
[15] Broadway, K. M., Modise, T., Jensen, R. V., & Scharf, B. E. (2014). Complete genome sequence of Salmonella enterica serovar Typhimurium VNP20009, a strain engineered for tumor targeting. Journal of biotechnology, 192, 177-178.
[16] Bumann, D., & Valdivia, R. H. (2007). Identification of host-induced pathogen genes by differential fluorescence induction reporter systems. Nature protocols, 2(4), 770-777.
[17] Laha, D., Grant, R., Mishra, P., & Nilubol, N. (2021). The Role of Tumor Necrosis Factor in Manipulating the Immunological Response of Tumor Microenvironment. Frontiers in Immunology, 12, 1415.
[18] Ma, J., Zhang, Y. G., Xia, Y., & Sun, J. (2010). The inflammatory cytokine tumor necrosis factor modulates the expression of Salmonella typhimurium effector proteins. Journal of Inflammation, 7(1), 1-14.
[19] Wang, J. X., Choi, S. Y., Niu, X., Kang, N., Xue, H., Killam, J., & Wang, Y. (2020). Lactic acid and an acidic tumor microenvironment suppress anticancer immunity. International Journal of Molecular Sciences, 21(21), 8363
[20] Bai, R., Lv, Z., Xu, D., & Cui, J. (2020). Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomarker Research, 8(1), 1-17
[21] Cronin, M., Akin, A. R., Collins, S. A., Meganck, J., Kim, J. B., Baban, C. K., & Tangney, M. (2012). High resolution in vivo bioluminescent imaging for the study of bacterial tumour targeting. PloS one, 7(1), e30940.
[22] Brodl, E., Winkler, A., & Macheroux, P. (2018). Molecular mechanisms of bacterial bioluminescence. Computational and structural biotechnology journal, 16, 551-564.
[23] Wille, T., Blank, K., Schmidt, C., Vogt, V., & Gerlach, R. G. (2012). Gaussia princeps luciferase as a reporter for transcriptional activity, protein secretion, and protein-protein interactions in Salmonella enterica serovar typhimurium. Applied and environmental microbiology, 78(1), 250-257.
[24] Welter, M., & Rieger, H. (2016). Computer simulations of the tumor vasculature: applications to interstitial fluid flow, drug delivery, and oxygen supply. Systems Biology of Tumor Microenvironment, 31-72.
[25] Fredrich, T., Rieger, H., Chignola, R., & Milotti, E. (2019). Fine-grained simulations of the microenvironment of vascularized tumours. Scientific reports, 9(1), 1-12.
