
Proposed Implementation
Introduction
When starting the development of our product, one of our first challenges was to understand where it would be applicable in the real world, and who our end users would be. We knew early on that our project is generic and purposely not focused on one specific solution. It can be implemented in many different fields involving bacterial communities, whether they stay in a lab or in a natural microbiome.
We started exploring the leading fields in which microbiome engineering is taking place, as well as fields we felt our product could be useful. After doing our own initial research, we further expanded on these ideas by speaking with many researchers in the field, as well as biotech industry leaders. These people helped shaped the possible implementation for our product, which is summarized below.
Much of the questions we asked throughout our thinking process, and how we implemented those into our project, are summarized in the Integrated Human Practicessection.
Agriculture
Food security is being threatened more than ever by the rapid changes our planet is facing, including a booming population, climate change, depletion of natural resources, and more [11]. Now more than ever, farmers are looking for more sustainable farming methods, while scientists are looking how they can increase food availability [1]. Increasing yields by improving plant growth is just one way to do so, and synthetic biologists are looking at ways to both modify plants as well as the organisms which affect their growth so significantly. While chemical fertilizers can be used to increase yield, in many cases they also cause serious environmental and health concerns [28]. Therefore, many efforts have been made to shy away from chemicals and move towards biological fertilizers.
Genetic modification of organisms (GMO) is one such effective proposed bio-fertilizer. While plants have been bred and modified for many generations, new technology now allows for quicker, more exact, and more significant gene editing, increasing the potential of how much we can affect plant growth. However, genetic engineering of food comes with a lot of criticism, as people and organization fear issues such as potential toxicity, possible gene transfer to other species, pollution, and more [27]. Therefore, regulations on food GMOs are particularly tight around the world, and progress in the field is mostly limited to the lab.
Plants can be considered holobionts - or, a host together with the many other species living in or around it - as plants live in nearly constant symbiosis with microorganisms such as bacteria and mycorrhizal fungi. These microorganisms play an important role in the plant’s life and fitness [5], and form communities in the soil which interact with the roots and affect plant growth [23]. These microorganisms can also be manipulated by gene editing to increase growth in plants via many methods, such as increasing phosphorous availability, metabolite production, and more [3, 7, 13]. We propose using our software for selective and safe genetic modification of rhizosphere and soil microbiome at a much lower risk than plant modification.
Currently viable applications include utilization of soil samples in order to engineer bacterial-based enhancements for crops, while avoiding any form of plant genetic modification. Such enhancements include the engineering of soil-specific biofertilizers or biopesticides by examining the natural state and bacterial composition of the soil and providing “personalized medicine” for the field.

Figure 1: Using Communique in agriculture. Made with Biorender
This proposed agricultural engineering exceeds existing alternatives not only by offering a safe and eco-friendly solution, but by providing much higher lifetime value. The initial treatment could be maintained through periodic tweaks over time, forming a long-term relationship with the client. Such processes could be integrated into existing field care regimens in a highly personalized and specific manner, designed to fit the exact needs of farmers or agriculture companies.
Our vision is to innovate agrotech and reduce the use of harmful soil additives by integrating and optimizing the natural system and using cutting-edge microbial engineering.
Bioremediation of Oil Spills
Oil spills happen frequently and cause environmental and ecological issues. Whether by accident or deliberate, it’s estimated that thousands of tons of oil are lost in the water every year [8, 9]. These massive spills of hydrocarbons (such as petroleum and its products) cause long-term damage to the surrounding and distant marine ecosystems and wildlife, as well as off-shore ecosystems [17]. While the significant economic and environmental impacts are difficult to quantify [26], it is crucial to push forward development of preventative measures, as well as more effective clean-up solutions, in order to deal with this ongoing destruction.
One way to deal with oil spills is through bioremediation, a process in which microorganisms are used to degrade toxic chemicals and pollutants into components that can be safely returned to the environment [21]. The three main types of bioremediation used for petroleum spills include microbial remediation, phytoremediation (using living plants), and mycoremediation (fungi). These indigenous communities of microorganisms are able to break down
Other cleanup methods, such as chemical and manual removal, are more labor-intensive, expensive, and can cause chemical or mechanical damage [30].
Bacterial consortia are often found in the community of microorganisms which can degrade crude oil. These bacteria use these aromatic hydrocarbons as their only source of carbon and energy [21]. We can genetically engineer and take advantage of these communities in order to improve their efficiency and ability to degrade oil.
There are many studies which have already identified consortia, specific species, and key metabolic genes which participate in the bioremediation process [4, 6, 28, 22, 24]. Our system would both allow specific integration of new genes and pathways to a desired species, as well as increasing the biosafety of modifying natural communities by preventing horizontal gene transfer (HGT).
Human Microbiome
Background
The microbiota is composed of unique communities of microorganisms include bacteria, archaea, protists, fungi and viruses. These communities live on and within the human body, mainly in the gut, in the oral cavity, and on the skin. Quantitatively, for each human cell in the human body we have about 10 bacterial cells, and the total weight of all the microorganisms in an adults body is about 3 kg [14].
In the last decade, research in the field of microbiome is gaining momentum. Experiments show it has a huge potential, and an equally important role to genetics in influencing the differences between humans, health and disease states, cognition, emotions etc [16].
The initial composition of the microbiota is inherited from the mother to the newborn at birth and can be changed during the life. It is affected by a variety of internal and environmental factors such as: genetics, hygiene, nutrition, medication, living area, life partners, and stressful situations. Shifts in the composition of the microbiota are associated with the pathogenesis of obesity, diabetes, heart and liver diseases, chronic inflammatory bowel diseases, and gastrointestinal cancer, as well as autism and stress [10].
Through microbiome engineering, the unbalanced microbiome will be rebalanced, and the ecosystems will be restored to their balanced states or even improved, leading to enhanced phenotypes [10]. A breakthrough in this field is a fecal microbiota transplant (FMT), a process of transferring fecal bacteria and other microbes from a healthy individual into another individual, to rebuild the patient's microbiota. This method was proven as an effective treatment for Clostridioides difficile infection (CDI) [12].
By using our software tool, it will be possible to engineer the microbiome by causing a desired gene to be expressed in a specific population and through this changing the bacterial composition. Some examples for applications are presented below.
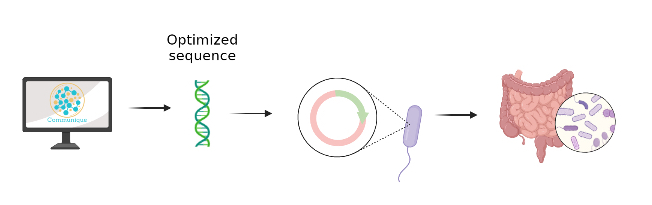
Figure 2: Implementation of Communique in the human microbiome. Made in Biorender.
Anti-cancer treatment [29]
The gut is bidirectionally connected to the nervous system through what is known as the “gut-brain axis” (GBA). As such, the gut serves as a complex interface between the resident bacteria in the gut microbiome and the rest of the human body, with the gut functioning as the communication gatekeeper between the host’s GBA and the resident bacteria in the gut microbiome.
Unsurprisingly (considering the extensive crosstalk between the host and its gut microbiota), certain microbes in the human gut microbiome have been shown to secrete certain molecules and metabolites that have been shown to help the host fight tumors and prevent tumorigenesis in several ways. In addition, it has also been shown that certain other types of bacteria secrete effectors that have pro-tumoral effects and promote cancer development.
Using our solution, it would be possible to selectively engineer the anti-tumoral bacteria to increase effector secretion and/or protect them from hostile factors reducing their population, thus improving their anti-cancer capabilities, and simultaneously engineer pro-tumoral bacteria to reduce secretion of pro-tumoral effectors and/or reduce their population — all in a safe and efficient manner.
Urea cycle disorders [15]
Urea cycle disorders (UCD) are inherited metabolic disturbances caused by deficiency in enzymes required to transfer nitrogen from ammonia to urea, which is created in the metabolism of proteins. Current treatment for UCDs focuses on dietary manipulations, ammonia-scavenging medication, and liver transplantation.
Recently, administration of an engineered ammonia hyperconsuming strain of Lactobacillus plantarum was found to reduce ammonia levels and mortality in rodents with hyperammonemia (a form of UCD). In addition, an orally-delivered E. coli probiotic which has been engineered to convert ammonia into arginine was found to effectively reduce systemic ammonia levels and improve survivability in hyperammonemia mouse models.
Using our solution, the risk of lateral gene transfer that arises from the incorporation of foreign engineered bacteria into the microbiome would be eliminated, as it would be possible to selectively engineer bacteria that are already present in the microbiome to reduce ammonia levels, thus reducing the disturbance to the delicate microbiome, as no external bacteria would be introduced.
Food industry

Figure 3: Implementation of Communique in the food industry. Made in Biorender.
Communities of microorganisms are a key part of fermentation, stability, nutrition, and deterioration of food [2]. New technologies in the previous years have allowed deeper analysis and understanding of these microbial landscapes and interactions [25]. Our technology will allow the industry to take advantage of this knowledge and tailor these bacteria to their benefit, whether it be lengthening shelf-life or increasing fermentation efficiency. It could eliminate production-line bottlenecks of natural bacterial consortiums, by specifically tweaking relevant genes and enzymes involved in the process.
Food enzymes, used widely in the food industry, are often extracted from microorganisms such as bacteria. It is becoming more common to genetically modify these bacteria to increase production and efficiency. Our tool could further increase productivity by taking advantage of synergistic effects of bacteria in a consortium, as well as increase the safety of genetically modifying organisms by preventing horizontal gene transfer. This could provide useful in overcoming regulations that are associated with use of genetically modified organisms (GMO’s) in the food industry in general.
One example is the process of yogurt production. The main cultures in yogurt are Lactobacillus bulgaricus and Streptococcus thermophilus [20]. The function of the starter cultures is to ferment lactose (milk sugar) to produce lactic acid. The increase in lactic acid produces the yogurt’s flavor, decreases pH and causes the milk to clot, or form the soft gel that is characteristic of yogurt.
Other bacterial cultures, such as Lactobacillus acidophilus, Lactobacillus subsp. casei, and Bifidobacteria may be added to yogurt as probiotic cultures [20]. Probiotic cultures benefit human health by improving lactose digestion, gastrointestinal function, and stimulating the immune system [19].Using Communique, we can optimize bacterial activity in these processes, thus increasing the lactic acid production and the probiotic cultures activity [19].
Biosensing
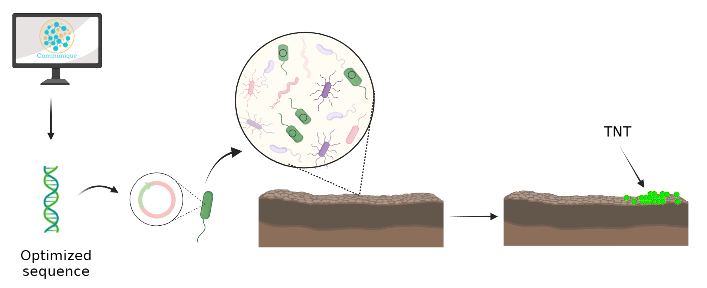
Figure 4: Implementation of Communique for Biosensing. Made in Biorender.
Bacteria can be engineered to act as biosensors for certain chemical and biological molecules. Chemicals and pollutants break down in soil, releasing small molecules that can be detected by bacteria, which in are coupled to a reporter gene that can give a detectable response. In the past, these bacterial biosensors have been used to detect a range of analytes, or substances. For example, they’ve been used for detecting toxic metals like arsenic, mercury, and lead, nitrate in the rhizosphere, and pollutants in the environment [32, 34, 35, 37]. Another prominent example is the detection of landmines by the way of sensing trinitrotoluene (TNT) and its degradation product, 2,3-dinitrotoluene (DNT), which is considered even more volatile and easy to sense [36]. Landmines are considered a concern around the world, and especially here in Israel, where there are an estimated 260,000 landmines [33]. Research has already shown success in using bacterial biosensors to accurately detect landmines via DNT and TNT [36].
Our conversations with Yuval Dorfan, director of the Israeli SynBio Institute (ISI) and Liam Kimmel helped us realize our software’s ability to increase the possibilities and safety for the bacterial biosensing software. This particular biosensing solution requires releasing bacteria into the soil, which poses the same biosafety risks associated with horizontal gene transfer. We propose two different strategies to use our software in landmine biosensing that we believe could greatly reduce the associated risks. First, bacteria could be engineered using our optimized plasmids and then sprayed into the soil for detection. The optimization would be strong enough to ensure that, should HGT occur, other species would not be able to express the optimized gene. Second, the engineered plasmid itself could be sprayed into the soil rather than bacteria themselves, with the detectable gene optimized for a species that already exists in the soil. This would, however, require engineering both a sensor and a reporter gene, or utilizing one which exists in the species already.
In addition to using bacteria as biosensors, our plasmid itself could act as a powerful biosensor to detect bacteria. Different techniques have been used in the past to detect harmful bacterial strains, whether they be pathogenic bacteria in water sources or bacterial contaminants in beer fermentation [31, 38]. Our plasmid could be used to express reporter genes optimally and specifically in the target strains, to detect presence and quantity.
Research
Several of the researchers we spoke to expressed interest in using our software in their own laboratories and experiments. The uses range from more efficient transformation into bacterial strains, to observing functionality and relationships within microbiomes. More about this can be read on our Integrated HP page.
References
- B. Lenaerts, B. C. Y. Collard, and M. Demont, “Review: Improving global food security through Accelerated Plant Breeding,” Plant Science, vol. 287, p. 110207, 2019.
- E. Caplice, “Food fermentations: Role of microorganisms in food production and preservation,” International Journal of Food Microbiology, vol. 50, no. 1-2, pp. 131–149, 1999.
- E. Malusá, L. Sas-Paszt, and J. Ciesielska, “Technologies for beneficial microorganisms Inocula used as Biofertilizers,” The Scientific World Journal, vol. 2012, pp. 1–12, 2012.
- E. S. Gilbert, A. W. Walker, and J. D. Keasling, “A constructed Microbial Consortium for biodegradation of the Organophosphorus Insecticide Parathion,” Applied Microbiology and Biotechnology, vol. 61, no. 1, pp. 77–81, 2003.
- E. T. Kiers and M. G. Heijden, “Mutualistic stability in the arbuscular mycorrhizal symbiosis: Exploring hypotheses of evolutionary cooperation,” Ecology, vol. 87, no. 7, pp. 1627–1636, 2006.
- H. Liu, J. Yao, Z. Yuan, Y. Shang, H. Chen, F. Wang, K. Masakorala, C. Yu, M. Cai, R. E. Blake, and M. M. F. Choi, “Isolation and characterization of crude-oil-degrading bacteria from oil-water mixture in Dagang oilfield, China,” International Biodeterioration & Biodegradation, vol. 87, pp. 52–59, 2014.
- H. Rodriguez, T. Gonzalez, I. Goire, and Y. Bashan, “Gluconic acid production and phosphate solubilization by the plant growth-promoting bacterium azospirillum spp..,” Naturwissenschaften, vol. 91, no. 11, pp. 552–555, 2004.
- H. Singh, N. Bhardwaj, S. K. Arya, and M. Khatri, “Environmental impacts of oil spills and their remediation by magnetic nanomaterials,” Environmental Nanotechnology, Monitoring & Management, vol. 14, p. 100305, 2020.
- J. Chen, W. Zhang, Z. Wan, S. Li, T. Huang, and Y. Fei, “Oil spills from Global Tankers: Status Review and future governance,” Journal of Cleaner Production, vol. 227, pp. 20–32, 2019.
- J. L. Foo, H. Ling, Y. S. Lee, and M. W. Chang, “Microbiome engineering: Current applications and its future,” Biotechnology Journal, vol. 12, no. 3, p. 1600099, 2017.
- J. Mockshell and M. E. Villarino, “Agroecological intensification: Potential and limitations to achieving food security and Sustainability,” Encyclopedia of Food Security and Sustainability, pp. 64–70, 2019.
- J. S. Bakken, T. Borody, L. J. Brandt, J. V. Brill, D. C. Demarco, M. A. Franzos, C. Kelly, A. Khoruts, T. Louie, L. P. Martinelli, T. A. Moore, G. Russell, and C. Surawicz, “Treating clostridium difficile infection with fecal microbiota transplantation,” Clinical Gastroenterology and Hepatology, vol. 9, no. 12, pp. 1044–1049, 2011.
- J. Zhu, M. Li, and M. Whelan, “Phosphorus activators contribute to legacy phosphorus availability in agricultural soils: A Review,” Science of The Total Environment, vol. 612, pp. 522–537, 2018.
- L. K. Ursell, J. L. Metcalf, L. W. Parfrey, and R. Knight, “Defining the human microbiome,” Nutrition Reviews, vol. 70, 2012.
- L. R. Soria, N. Ah Mew, and N. Brunetti-Pierri, “Progress and challenges in development of new therapies for urea cycle disorders,” Human Molecular Genetics, vol. 28, no. R1, 2019.
- M. A. Malla, A. Dubey, A. Kumar, S. Yadav, A. Hashem, and E. F. Abd_Allah, “Exploring the human microbiome: The potential future role of next-generation sequencing in disease diagnosis and treatment,” Frontiers in Immunology, vol. 9, 2019.
- M. G. Barron, D. N. Vivian, R. A. Heintz, and U. H. Yim, “Long-term ecological impacts from oil spills: Comparison of Exxon Valdez, Hebei spirit, and Deepwater Horizon,” Environmental Science & Technology, vol. 54, no. 11, pp. 6456–6467, 2020.
- M. Nitschke and G. M. Pastore, “Production and properties of a surfactant obtained from bacillus subtilis grown on cassava wastewater,” Bioresource Technology, vol. 97, no. 2, pp. 336–341, 2006.
- “Milk facts,” Milk and Human Health | MilkFacts.info. [Online]. Available: http://www.milkfacts.info/Nutrition%20Facts/Milk%20and%20Human%20Health.htm#Probiotics. [Accessed: 18-Oct-2021].
- O. Adolfsson, S. N. Meydani, and R. M. Russell, “Yogurt and gut function,” The American Journal of Clinical Nutrition, vol. 80, no. 2, pp. 245–256, 2004.
- O. B. Ojuederie and O. O. Babalola, “Bioremediation of heavy metals and hydrocarbon polluted environments: A Review,” 2017.
- P. Dvořák, P. I. Nikel, J. Damborský, and V. de Lorenzo, “Bioremediation 3 . 0 : Engineering pollutant-removing bacteria in the times of systemic biology,” Biotechnology Advances, vol. 35, no. 7, pp. 845–866, 2017.
- P. Vandenkoornhuyse, A. Quaiser, M. Duhamel, A. Le Van, and A. Dufresne, “The importance of the microbiome of the plant holobiont,” New Phytologist, vol. 206, no. 4, pp. 1196–1206, 2015.
- Q. Wang, X. Fang, B. Bai, X. Liang, P. J. Shuler, W. A. Goddard, and Y. Tang, “Engineering bacteria for production of RHAMNOLIPID as an agent for enhanced oil recovery,” Biotechnology and Bioengineering, vol. 98, no. 4, pp. 842–853, 2007.
- Q. Yu, Y. Li, B. Wu, W. Hu, M. He, and G. Hu, “Novel mutagenesis and screening technologies for food microorganisms: Advances and prospects,” Applied Microbiology and Biotechnology, vol. 104, no. 4, pp. 1517–1531, 2020.
- S. E. Chang, J. Stone, K. Demes, and M. Piscitelli, “Consequences of oil spills: A review and framework for informing planning,” Ecology and Society, vol. 19, no. 2, 2014.
- S. G. Uzogara, “The impact of genetic modification of human foods in the 21st Century,” Biotechnology Advances, vol. 18, no. 3, pp. 179–206, 2000.
- S. Savci, “Investigation of effect of chemical fertilizers on environment,” APCBEE Procedia, vol. 1, pp. 287–292, 2012.
- S. Vivarelli, R. Salemi, S. Candido, L. Falzone, M. Santagati, S. Stefani, F. Torino, G. L. Banna, G. Tonini, and M. Libra, “Gut microbiota and cancer: From pathogenesis to therapy,” Cancers, vol. 11, no. 1, p. 38, 2019.
- X. M. E. Refugio RV, “Microorganisms metabolism during bioremediation of oil contaminated soils,” Journal of Bioremediation & Biodegradation, vol. 07, no. 02, 2016.
- A. G. Prest, M. K. Winson, J. R. M. Hammond, and G. S. A. B. Stewart, “The construction and application of a LUX ‐based nitrate biosensor,” Letters in Applied Microbiology, vol. 24, no. 5, pp. 355–360, 1997.
- A. Hynninen and M. Virta, “Whole-cell bioreporters for the detection of bioavailable metals,” Whole Cell Sensing System II, pp. 31–63, 2009.
- J. S. Kreniske, A. Harris, and W. Safadi, “Landmines in the Golan Heights: A patient's perspective,” Case Reports, vol. 2014, no. aug24 1, 2014.
- K. M. DeAngelis, P. Ji, M. K. Firestone, and S. E. Lindow, “Two novel bacterial biosensors for detection of nitrate availability in the rhizosphere,” Applied and Environmental Microbiology, vol. 71, no. 12, pp. 8537–8547, 2005.
- R. Tecon and J. Van der Meer, “Bacterial biosensors for measuring availability of environmental pollutants,” Sensors, vol. 8, no. 7, pp. 4062–4080, 2008.
- S. Belkin, S. Yagur-Kroll, Y. Kabessa, V. Korouma, T. Septon, Y. Anati, C. Zohar-Perez, Z. Rabinovitz, A. Nussinovitch, and A. J. Agranat, “Remote detection of buried landmines using a bacterial sensor,” Nature Biotechnology, vol. 35, no. 4, pp. 308–310, 2017.
- S. Ramanathan, M. Ensor, and S. Daunert, “Bacterial biosensors for monitoring toxic metals,” Trends in Biotechnology, vol. 15, no. 12, pp. 500–506, 1997.
- Y. Wu, C.-W. Wang, D. Wang, and N. Wei, “A whole-cell biosensor for point-of-care detection of waterborne bacterial pathogens,” ACS Synthetic Biology, vol. 10, no. 2, pp. 333–344, 2021.












