
Project Description
The Problem
In the US, CRC is the fourth most common cancer, with female breast, prostate, and lung and bronchus cancer ahead. In 2018, 141,000 new CRC cases were reported, and 52,000 people died of CRC. In the US, 1 in 23 men and 1 and 25 women will be diagnosed with CRC in their lifetime1. The 5 years survival rate for CRC is 64%, but early detection can raise that rate to as high as 90%2. There are many CRC detection methods available, with the most common being colonoscopies. However, ~¼ of adults are not getting screened as recommended for CRC, likely due to inability to access necessary tests, inability to afford tests, and the invasive nature of some tests (colonoscopy, sigmoidoscopy)3.
Inspiration
To address this issue, we wanted to design a system that could detect CRC through producing a change in stool color. Current alternative detection methods, such as ColoGuard, were examined as a starting point and to ensure a novel system. We explored two potential methods for detection: FimH/OxyR mediated response and quorum sensing; Long-term versus short-term detection was also explored.
Further investigation and conversations with microbiology experts led us to settle on short-term detection through FimH/OxyR mediated response.
The Solution
We propose Colon Detective as a at home colon cancer detection system which aims to detect colon cancer cells by producing a color change in stool. The user will ingest a probiotic, which will contain engineered E. coli cells. These engineered cells will express antitumor antibodies that will specifically bind to colon cancer cells. The binding of these antibodies will activate OxyR, a transcriptional activator. This activation induces the production of a beta-galactosidase reporter which will cause the stool to become blue when exposed to a compound known as x-gal. Following ingestion of the probiotic, application of x-gal will cause a color change in stool if cancerous cells are detected.
FimH Biniding
The second main aspect of the system is FimH binding. FimH is the capping protein of the bacterial fimbriae. Our goal is to express the FimH-SNAP fusion protein on the surface of our bacteria.

The SNAP protein would allow different antibodies to be covalently bonded to the FimH protein via O6-benzylguanine on the antibody. Since SNAP-tag forms a covalent linkage with O6-benzylguanine derivatives, antibodies conjugated to guanine would be bounded to the FimH. We plan to use this method to localize the bacteria around the cancer cells with specific antibodies for CRC. In order to express our protein of interest, we created a plasmid which expresses FimH-SNAP with the pRSF-Duet backbone.

OxyR Induction
The first main aspect of the system is OxyR activation. OxyR is a transcription factor that, when active, induces the production of a beta-galactosidase reporter. We isolated three known OxyR regulator genes that can be modified to produce lacZ: katG, dps, and grxA.
OxyR presents two strategies that are sensitive to the cancer cell environment. First, the cancer cell environment is hypoxic and OxyR’s transcriptional activity is directly regulated by H2O2. Secondly, it has been shown that attachment of FimH to surfaces stimulates expression mediated by OxyR. In order to test hypoxic induction of OxyR, we modelled our experiments based on Rodríguez-Rojas et. al. (2020) to measure how H2O2 concentration impacts bacterial growth.

LacZ Production
The third main aspect of our project is LacZ production. OxyR activation leads to upregulation of RNA Polymerase, transcribing the genes that have been modified to produce lacZα.

Once lacZα is produced, it will spontaneously reassemble with lacZΩ and produce β-galactosidase.
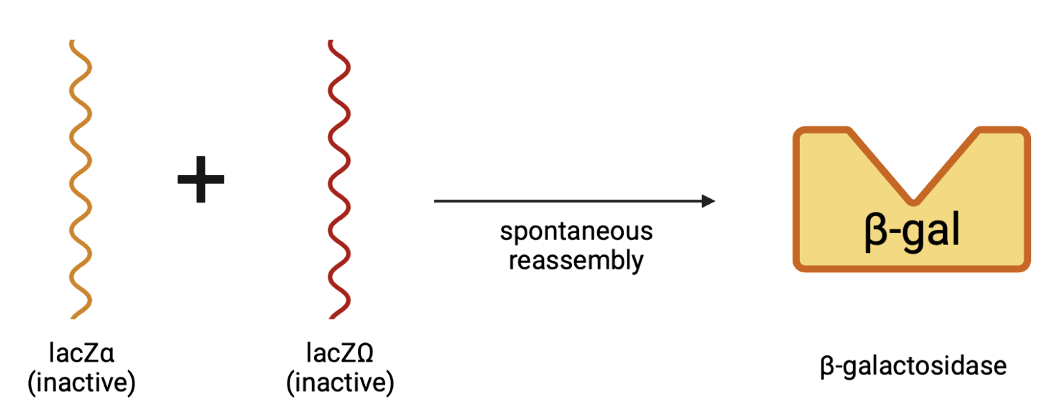
Following the production of our β-galactosidase, the cells will be exposed to a compound containing lysis buffer and x-gal. In the presence of β-gal, X-gal will turn blue, which will be seen in the stool to indicate detection of cancer cells

References
- USCS Data Visualizations https://gis.cdc.gov/Cancer/USCS/#/AtAGlance/.
- American Cancer Society. Colorectal Cancer Screening Test Use* (%), Adults 50 Years and Older by State, 2018; 2020.
- Use of Colorectal Cancer Screening Tests (2018 Behavioral Risk Factor Surveillance System) https://www.cdc.gov/cancer/colorectal/statistics/use-screening-tests-BRFSS.htm.

