Results


At the beginning of the experiment, we phosphorylated the single strand of DNA, that is, ATP provided the phosphate group, and PNK enzyme added phosphoric acid to the 5 'end of the single strand for subsequent loop formation.
Next, we added the T4 ligase to make the long single strand of DNA into a loop, and in this process, we cycled T1 and S2 in different ways. T1 uses its own hairpin structure to fix the two ends that need to be looped, and then lets T4 ligase repair the gap to make it looped. For S2, we add the auxiliary splint chain to fix both ends of the chain and increase the probability of loop formation.
At the same time, we also tried to use two short chains to connect a single ring structure, the main purpose of this is to reduce the economic cost. The single strand that was not connected to the loop and the splint strand were removed by ExoI after enzyme digestion. Thus, we have T1 and S2 monocycles with almost no impurities.
Hint: the abbreviation P stands for phosphorylation and the word C stands for cyclized and d stands for digested.
Long chain connection:


Short chain connection:
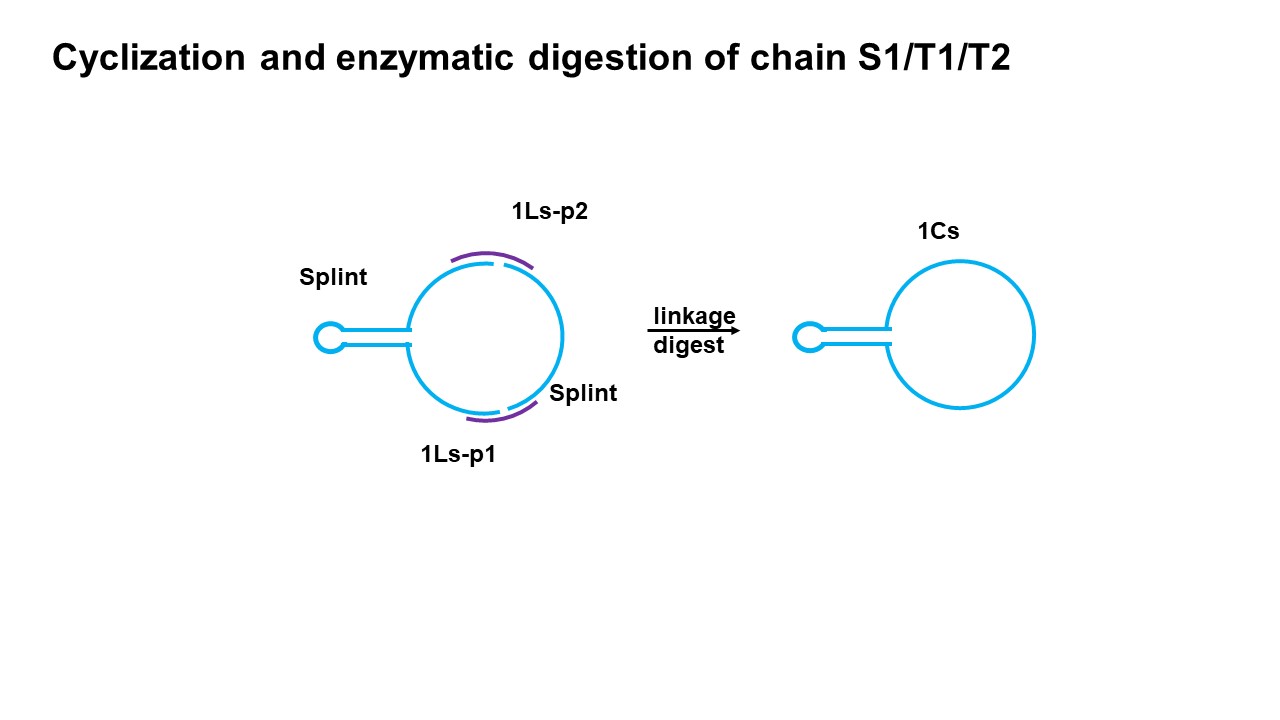







In order to better obtain the double ring structure, we adopt the method of two connected segments. The so-called two-segment link refers to a single ring and two short chains connected to form a double ring structure. The results show that this method can get the desired structure. In this step, we first put T1 single ring and S1P2 in the same system, and then put S1P1 in the other system, after annealing and denaturing together, add enough enzyme to S1P1 and mix the two systems quickly. Finally, put it into the appropriate temperature and react for 2 hours. Similarly, short chains T2P1 and T2P2 were added to S2 in the same way to form a double-ring structure.



After that, double ring enzymatic digestion was started, which was similar to single ring enzymatic digestion. In order to improve the purity of the product, exoIII with better effect was added, and the reaction was carried out at 37℃ for two hours, and the double ring structure was obtained.
It is speculated that Lk7 (δ LK-1) was prepared from channel 8, which may contain a small amount of Lk6 (δ LK-2) and Lk5 (δ LK-3). Lane 14 also has a small amount of Lk7. With the presence of topological limiting forces, Lk7 is easy to form.






In the end we transcribe.
In the transcription results without inducers, it can be seen that APP sequence and topological isomers with cross structure can promote transcription under experimental conditions, which may be because z-DNA structure is easy to form in the presence of high concentration of magnesium ions, and cross structure is opened at the same time. Of course, there is another situation, that is, a bubble structure is formed in the double ring structure, which makes the enzyme easy to bind to the bubble structure, thus making the initiation of transcription easier, what’s improving the efficiency of transcription.


From the results of adding Z22 protein and ZBP-1 protein for transcription, it can be seen that the added Z22 protein and ZBP-1 protein can inhibit gene transcription, and the inhibition effect of ZBP-1 is stronger than that of Z22. Although there is a certain gap between this and the expected experimental results, we will continue to improve our experiment, adjust the experimental conditions, To further explore the effects of left and right helix and cross structure on gene expression.






