
Parts
a) BBa_K39570000
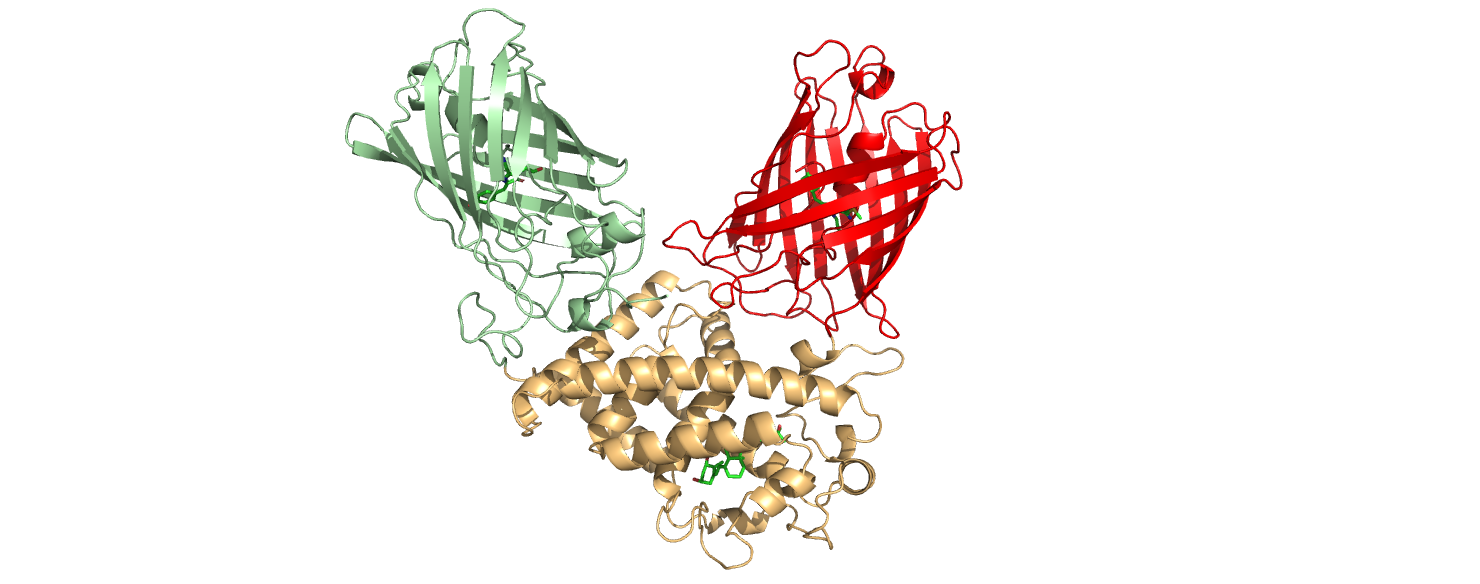
This part is an improvement of the part BBa_K3515011, designed by
Queen’s University iGEM in 2020. It consists of the ligand binding
domain of vitamin D binding receptor protein (VDR) from Homo
sapiens, as well as mNeonGreen (BBa_K1761003) and mCherry
(BBa_J06504) fluorophores linked to the N and C
terminal ends of the VDR LBD, respectively.
The improvements of this part include its codon optimization for
expression in Escherichia coli, the removal of a stop codon in the
middle of the sequence, the addition of a His-tag, and the removal
of restriction sites to ensure biobrick compatibility.
Vitamin D receptor binding protein interacts with
1,25-dihydroxyvitamin D to produce a conformational change.
Biologically, this process is used to start the cycle of protein
dimerization so that a protein heterodimer can bind to an enhancer
sequence and incite a transcriptional rate change. With regards to
the BBa_K3957000 construct, this conformational change results in
the movement of the terminal mNeonGreen and mCherry fluorophores.
mNeonGreen and mCherry are fluorescent proteins; They absorb light
at specific wavelengths, and emit light at different, specific
wavelengths. When paired together, the mNeonGreen and mCherry
fluorophores exhibit a phenomenon called FRET. FRET, or fluorescence
resonance energy transfer, is a mechanism in which the energy
absorbed by one fluorescent molecule is transferred to a secondary
fluorescent molecule of a different type while in close proximity,
resulting in the emittance of a different wavelength of light.The
difference between the emittance of the secondary fluorescent
molecule before and after FRET induction may be used as a reporter
system. In the case of BBa_K3957000, the primary (light absorbing)
fluorophore is mNeonGreen, and the secondary (light emitting)
fluorophore is mCherry. As mentioned, since FRET signal strength is
based on the proximity of the two fluorophores, conformational
change in the ligand binding domain of VDR can create a change in
the signal of FRET. Therefore, this part was used to report the
binding of 1,25-dihydroxyvitamin D in our project.
b) BBa_K3957002
This part is a standalone construct built by our team. It consists
solely of an optimized coding region for the human 1,
25-alpha-hydroxylase enzymatic protein (referred to as OHase).
OHase functions to catalyze the addition of a hydroxyl group to the
1-C position of 25-hydroxyvitamin D, converting it to the
biologically active form of vitamin D (1,25-dihydroxyvitamin D).
This enzyme is useful for studying vitamin D in a biological
environment, as the inactive form (25-hydroxyvitamin D) is found in
abundance relative to the active form.
Part BBa_K3957002 was optimized for expression in Escherichia coli
as the 1, 25-alpha-hydroxylase sequence originated from Homo
sapiens.
This part was planned to be used in our project as a means of
methodologically simulating the conversion of 25-hydroxyvitamin D to
1,25-dihydroxyvitamin D, as it is an integral step when measuring
vitamin D levels in biological systems. However, due to time
constraints, the part was not utilized in our experiments.
