Overview
As a result of the COVID-19 pandemic, most of the work done by the team was online. The team thus had to carry out all of the computational work on local systems.
We plan on finding an enzyme that leads to degradation of gossypol without changing other nutritional characteristics. This will allow us to utilize cottonseed meal as feed for small non-ruminants like sheep, goats, etc. as well as present a viable option for human consumption due to cottonseed being rich in proteins and vitamins. For more details on the project, you can refer to our Engineering page.
This enzyme can change the functional nature of gossypol by neutralizing its toxicity. Ultimately, a cost effective process will lead to the production of non-toxic CSM and cottonseed oil and also benefit several industries as well.
On this page we will be covering the enzyme selection process. The Dry Lab Workflow was divided into 3 distinct sections:
In-silico Screening
This section focuses on trying to identify the potential enzymes that are capable of degrading gossypol. This section is further divided into 3 subsections :
1. Initial Shortlisting
Gossypol is a polyphenolic molecule present all throughout the cotton plant. Gossypol is predominantly found in the cell wall of the plant cell and is closely associated with lignin within the cell wall. These characteristics were taken into account in order to carry out a literature search, i.e., we started searching for enzymes that act on naphthalene rings, fused furan rings and/or aldehydes. Also the fact that gossypol is present within the lignin of the cell wall was accounted for and enzymes attacking lignin were also evaluated. We placed special emphasis on enzymes that satisfied both criteria, i.e., attacking one of the functional groups and attacking lignin.
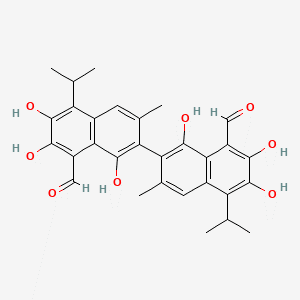
Fig 1: Gossypol[1]
After extensive literature review, we identified the following enzymes that could be used to degrade gossypol. All the enzymes, having different structural and chemical properties, bind to ligands differently, hence, all the characteristics of the enzyme were noted and selected accordingly.
- Lignin peroxidases[2]
- Manganese peroxidases [2]
- Laccases [3]
- Cytochrome P450[4]
- Monooxygenases[4]
- Salicylaldehyde dehydrogenases[5]
- Phenol hydrolases [6]
Along with going through the existing literature, we decided to use computational methods in order to search for the enzymes that could be used in our project.
We used a web server known as PharmMapper that identifies potential target candidates for the given probe small molecules. But it was decided not to utilize this server for the screening process as it follows a reverse pharmacophore mapping strategy which doesn't align with our project.
Download the results by clicking this button: Download Pharmmapper results
STITCH (Search Tool for Interaction of Chemicals) incorporates data and information regarding chemical interactions between molecules from different metabolic pathways, binding interactions and drug-target interactions. Databases like PubChem, ChEBI, ChemDB are utilized to find interactions between the given molecule and all other molecules in the database[7]. When we ran gossypol on the server, it returned only molecules produced by Homo sapiens, whereas we were looking for molecules produced by bacteria and/or yeast since production of these will be more feasible through Pichia pastoris. Hence, we decided to reject these results and continued seeking other methods.
Download obtained results by clicking this button: Download STITCH results

Fig 2: STITCH Result for Gossypol
2. Docking
CB-Dock is a free online server that enables its users to identify the binding sites of a protein. The server asks for a protein molecule as PDB file as well as the ligand molecule file and then applies blind docking technique and utilizes curvature-based cavity detection approach to recognize the top 5-10 binding sites as well as calculating the centers and sizes.The docking box size is determined on the basis of the ligand provided and the docking is performed on AutoDock Vina[8].

Fig 3: CB-Dock Result for 3FU8 with Gossypol
Here is the link for the molecules that were screened on the CB-dock server Download
TOP 5 ENZYMES THAT WERE IDENTIFIED VIA CB-DOCK :
2IJ2 : Flavocytochrome P450-BM3
| Vina score | Cavity size | Center | Size | ||||
|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||
| -9 | 5516 | 63 | 13 | 51 | 31 | 35 | 33 |
| -8.7 | 11012 | 72 | -4 | 66 | 35 | 35 | 33 |
| -7.9 | 5669 | 45 | 30 | 39 | 34 | 35 | 31 |
| -7.9 | 149 | 56 | 3 | 69 | 24 | 24 | 24 |
| -7.9 | 140 | 33 | 13 | 36 | 24 | 24 | 24 |
3FU8 : Melanocarpus albomyces laccase
| Vina score | Cavity size | Center | Size | ||||
|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||
| -8.5 | 3308 | -9 | 30 | 19 | 24 | 24 | 24 |
| -8.4 | 1092 | -16 | 37 | 43 | 24 | 24 | 24 |
| -8 | 1321 | -3 | 23 | 42 | 24 | 24 | 24 |
| -7.8 | 117 | 2 | 39 | 39 | 24 | 24 | 24 |
| -7.7 | 590 | -14 | 44 | 22 | 24 | 24 | 24 |
1TQN : Human Microsomal P450 3A4
| Vina score | Cavity size | Center | Size | ||||
|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||
| -8.4 | 11268 | -25 | -18 | -9 | 31 | 35 | 31 |
| -8.1 | 772 | -27 | -25 | -31 | 24 | 24 | 24 |
| -7.4 | 258 | -32 | -18 | -8 | 24 | 24 | 24 |
| -7.4 | 240 | -9 | -25 | -18 | 24 | 24 | 24 |
| -7.4 | 231 | -4 | -29 | -25 | 24 | 24 | 24 |
6T1U : Cytochrome P450 reductase from Candida tropicalis
| Vina score | Cavity size | Center | Size | ||||
|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||
| -9.2 | 3386 | -30 | -6 | -18 | 24 | 24 | 24 |
| -8.9 | 1719 | 4 | -13 | -34 | 32 | 24 | 24 |
| -8.6 | 5929 | -18 | 11 | -64 | 30 | 35 | 24 |
| -8.4 | 181 | -22 | -21 | -53 | 24 | 24 | 24 |
| -8.3 | 3221 | -5 | -4 | -52 | 24 | 24 | 24 |
6HN8 : BM3 heme domain
| Vina score | Cavity size | Center | Size | ||||
|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||
| -9.7 | 3904 | 12 | 15 | -11 | 24 | 30 | 24 |
| -8.2 | 175 | -5 | 18 | 0 | 24 | 24 | 24 |
| -8 | 250 | -14 | 9 | 11 | 24 | 24 | 24 |
| -7.9 | 8077 | -10 | -7 | -3 | 35 | 35 | 35 |
| -7.6 | 4475 | -3 | -8 | 6 | 24 | 35 | 24 |
Note: Compared to regular docking, blind docking is less reliable and less stable as the docking space is usually too large to sufficiently sample using a limited number of random searches. Nevertheless, blind docking is particularly valuable for discovering unexpected interactions that may occur in unidentified binding modes.
The screening process included docking the enzyme with gossypol and checking the score using the software AutoDock v4.2. This is a major step in the screening process as the analysis of the protein-ligand interactions grants an insight into the stability of the resulting compound. After deep scrutiny of the various ligand conformations, we ranked the enzymes based on their docking scores and narrowed down on the following 2 enzymes :
2IJ2 : Flavocytochrome P450-BM3
2IJ2 is a 2 chain structure which functions as a fatty acid monooxygenase. It is known to catalyze hydroxylation of medium and long-chain fatty acids, and the reductase domain aids in electron transfer from NADP to cytochrome P450[9].

Fig: 2IJ2 Enzyme
- The best complex for 2IJ2 and gossypol has a binding energy of -10.55 kJ/mol
- Coordinates of Central Grid Point of Maps = (46.021, 19.896, 35.424)
- Minimum coordinates in grid = (33.532, 9.036, 24.564)
- Maximum coordinates in grid = (58.510, 30.756, 46.284)

Fig: 2IJ2-Gossypol Complex
Docking result for 2IJ2, in .txt format Download results
3FU8 : Melanocarpus albomyces laccase
Laccases are enzymes which oxidize an extensive variety of phenolics, along with other organic and even inorganic substrates using molecular oxygen as a terminal electron acceptor. 3FU8 is a laccase produced by Melanocarpus albomyces laccase crystals soaked with 2,6-dimethoxyphenol, a common laccase substrate[10].

Fig: 3FU8 Enzyme
- The best complex for 3FU8 and gossypol has a binding energy of -10.49 kJ/mol
- Coordinates of Central Grid Point of Maps = (-8.916, 29.730, 20.148)
- Minimum coordinates in grid = (-27.996, 17.010, 8.700)
- Maximum coordinates in grid = (10.164, 42.450, 31.596)

Fig: 3FU8-Gossypol Complex
Docking result for 3FU8, in .txt format Download results
We have done these experiments with limited computational resources in our local systems, since we did not have access to our laboratories. Due to the limited computational power, all of the mutations we carried out took a lot of time to be completed and we were unable to finish the mutation work. We plan to complete the experiments that follow once we get access to our workstation at institute, which would let us avail better computational resources and thus gain faster and more reliable results.
In-silico Mutations
Once the screening process is completed, in-silico mutations will be carried out as altering the structure of the finalized enzymes would be beneficial as mutations could help in improving binding of the enzymes with the ligand(i.e., gossypol) as well as improving the functionality of the mechanism.
Mutations could be carried out in 2 ways:
- Amino Acid Substitution
- Codon Optimization
As we will be looking for an enzyme which can perfectly bind to gossypol at its active sites, thus forming a compound which no longer exhibits the toxic side effects shown by gossypol. To do this, the amino acids of the shortlisted enzymes interacting with the ligand gossypol will be identified and studied. This will be done using the software Discovery Studio.
Discovery Studio is designed to help life science researchers by modelling molecular structures and analyzing them. It specialises in structure based design through tools for protein-protein docking, antibody design and fragment-based placement.

Fig 3: Mutated 3FU8 molecule being examined in Discovery Studio

Fig 4: All amino acids of 3FU8 interacting with gossypol assessed in Discovery Studio
Once these amino acids are identified in every shortlisted enzyme, they will be edited and replaced with each of the 19 other amino acids using PyMol 2.5.
Post this, UCSF Chimera will be used to minimize the energy of the edited enzymes which will enable us to achieve the most stable form of that enzyme.
UCSF Chimera is an open-source tool developed for interactive visualization and lets users download high quality images and movies. It is extensively used for measurement purposes such as calculating the distances and angles of atoms or the centroids and axes of molecules.

Fig 5: 3FU8 in UCSF Chimera

Fig 6: Energy Minimization process initiated in UCSF Chimera
Testing the stability of the edited enzyme will be done by analyzing Ramachandran's plot. This plot lets us determine the permitted torsional angles of the amino acids which further grants an insight into the structural changes brought about by the replaced amino acid. The intricacies of Ramachandran's plot can be found here:


Fig 8: Ramachandran plot for native 2IJ2 (left image)
and mutated 2IJ2 at ALA 264 to ARG. (right image).
Here, possible outliers in the ramachandran plot will be checked, which shall serve as a safety check for determining the accuracy and the reliability of the predicted structure. If most of the amino acids lie in the favored and allowed regions, the structure will be validated as an accurate prediction.
The favored, allowed and generously allowed areas are differentiated on the basis of the intensity of the gray color in the chart. Darker areas tend to show favored amino acid turn, and it progressively decreases in favorability. We will perform Ramachandran plot on UCLA-DOE LAB - SAVES v6.0. For more information visit this page.
Protein Modelling
- SWISS Model
- RaptorX
Protein Modelling is the process by which a bioinformatics tool can predict the structure of a molecule when the tool is provided with an amino acid sequence. High throughput parameters such as NMR Spectroscopy and X-ray crystallography are utilized to model the three dimensional structure of the given sequence.
1. SWISS Model
SWISS model is a free online server that predicts protein structure using homology modelling. Homology modelling, also known as comparative modelling, is based on the biological fact that when two sequences share high similarity/identity, their respective structures are also similar. The server takes a FASTA sequence of amino acids and then carries out a BLAST search for a closely related template in the PDB database. High query coverage, high percentage identity, high alignment score and low e-value are important features that are desirable for the template sequence. Once the template sequence is matched, the template sequence is modelled [11].

Fig 9: 3FU8 in SWISS Model
2. RAPTORX
RaptorX is developed by Xu group, excelling at tertiary and contact prediction for protein sequences without close homologs in the Protein Data Bank (PDB). RaptorX predicts protein secondary and tertiary structures, contact and distance map, solvent accessibility, disordered regions, functional annotation and binding sites[12].
Using this server, we will perform structural analysis for the point mutations that we would have caused in the protein's primary structure and also analyze the compatibility between the binding sites and the ligand binding.
Furthermore, we will check for the viability of the structure using the Ramachandran plot and also perform energy minimization on the predicted structure.

Fig 10: 2IJ2 in RaptorX
Final Result
Despite 2IJ2 (flavocytochrome P450-BM3) being a great candidate for the project, we decided to move ahead with laccase as our only enzyme for gossypol degradation. The main reason for this was that not much research work has been conducted on laccases for gossypol degradation. This would give novelty to the project. Also laccases are a widely known enzyme family which display versatility and would have a greater chance of being accepted for public use since many protocols already exist for storage, transportation and use of laccases on an industrial network.
Thus we decided to use 3FU8 (Melanocarpus albomyces laccase) as the enzyme for enzymatic Gossypol Degradation.
References:
-
[1] National Center for Biotechnology Information (2021).
PubChem Compound Summary for CID 3503, Gossypol. Retrieved October 17, 2021 from DOI: https://pubchem.ncbi.nlm.nih.gov/compound/Gossypol - [2] Fackler, K., Gradinger, C., Hinterstoisser, B., Messner, K., & Schwanninger, M. (2006). Lignin degradation by white rot fungi on spruce wood shavings during short-time solid-state fermentations monitored by near infrared spectroscopy. Enzyme and Microbial Technology, 39(7), 1476-1483. DOI: https://doi.org/10.1016/j.enzmictec.2006.03.043
-
[3] Wang L, Chen M, Luo X, Fan Y, Zheng Z, He Z, Yin R, Meng T, Xu S, Pan Y, Su J, Du J, Zhang L, Tian X, Tian
Y, Chen D, Ge H, Zhang N and Li P (2020) Intramolecular Annulation of Gossypol by Laccase to Produce Safe
Cottonseed Protein. Front. Chem. 8:583176.
DOI: https://doi.org/10.1016/S1872-2040(17)61045-4 - [4] Chen, C., Zhang, Y., Pi, W. et al. Optimization of the process parameters for reduction of gossypol levels in cottonseed meal by functional recombinant NADPH-cytochrome P450 reductase and cytochrome P450 CYP9A12 of Helicoverpa armigera. AMB Expr 9, 98 (2019). DOI: https://doi.org/10.1186/s13568-019-0823-4
-
[5] Jia, B., Jia, X., Hyun Kim, K. et al. Evolutionary, computational, and biochemical studies of the
salicylaldehyde dehydrogenases in the naphthalene degradation pathway. Sci Rep 7, 43489 (2017).
DOI: https://doi.org/10.1038/srep43489 - [6] Mohapatra Balaram, Phale Prashant S., Microbial Degradation of Naphthalene and Substituted Naphthalenes: Metabolic Diversity and Genomic Insight for Bioremediation, 2021. DOI: https://doi.org/10.3389/fbioe.2021.602445
- [7] Kuhn, M., von Mering, C., Campillos, M., Jensen, L. J., & Bork, P. (2008). STITCH: interaction networks of chemicals and proteins. Nucleic acids research, 36(Database issue), D684-D688. DOI: https://doi.org/10.1093/nar/gkm795
-
[8] Liu, Y., Grimm, M., Dai, Wt. et al. CB-Dock: a web server for cavity detection-guided protein-ligand blind
docking. Acta Pharmacol Sin 41, 138-144 (2020).
DOI: https://doi.org/10.1038/s41401-019-0228-6 - [9] https://proteopedia.org/wiki/index.php/2ij2
- [10] Kallio, J. P., Auer, S., Jänis, J., Andberg, M., Kruus, K., Rouvinen, J., Hakulinen, N. (2009). Structure-Function Studies of a Melanocarpus albomyces Laccase Suggest a Pathway for Oxidation of Phenolic Compounds. Journal of Molecular Biology, 392(4), 895-909. DOI: https://doi:10.1016/j.jmb.2009.06.053
- [11] Andrew Waterhouse, Martino Bertoni, Stefan Bienert, Gabriel Studer, Gerardo Tauriello, Rafal Gumienny, Florian T Heer, Tjaart A P de Beer, Christine Rempfer, Lorenza Bordoli, Rosalba Lepore, Torsten Schwede, SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Research, Volume 46, Issue W1, 2 July 2018, Pages W296-W303. DOI: https://doi.org/10.1093/nar/gky427
- [12] Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012 Jul 19;7(8):1511-22. DOI: https://doi.org/10.1038/nprot.2012.085 PMID: 22814390; PMCID: PMC4730388.
