The methane pillar of Cattlelyst aims to engineer the conversion of methane into biomass, similar to what happens in natural methanotrophs such as in M. capsulatus . The heterologous expression of methane monooxygenase, converting methane to methanol, remains a bottleneck in synthetic methanotrophy [1]. In this project both the membrane-bound particulate, as well as the soluble methane monooxygenase were explored. The expression of a particulate methane monooxygenase was suggested successful, yet no enzyme activity in vivo nor in vitro was detected. Additionally, the growth of an C1-auxotrophic E, coli C1Saux strain expressing methanol dehydrogenase from a plasmid was noted on formate and methanol. . Additionally, the methylotrophy of the synthetic E. coli SM1 strain was verified. This completed the conversion pathway of methanol into biomass and carbon dioxide, an important aspect of the methane pillar within the Cattlelyst project.

Introduction
Methane is a potent greenhouse gas. Via bioremediation, this gas can be neutralized into either the less potent carbon dioxide or used to accumulate biomass. Natural methanotrophs, such as the obligate methanotroph Methylococcus capsulatus, consume this methane and, via intermediary compounds, convert it into biomass for growth. However, the growing conditions of our biofilter are not favorable for these bacteria, as they are generally slightly thermophilic (45°C). Moreover, their slow growth [2] severely limits their genetic accessibility. In natural methanotrophs, the first step in the conversion of methane is facilitated by methane monooxygenase (MMO). M. capsulatus (Bath) is an unique species in this sense, as it contains both the sensitive, membrane-bound, particulate methane monooxygenase (pMMO) and the soluble methane monooxygenase (sMMO). The reaction mechanisms of pMMO and sMMO are as shown in Figure 2 and Figure 3, respectively.


The second step in the oxidation of methane and the final step before biomass can be assimilated is performed in M. capsulatus by methanol dehydrogenase (Mdh). This enzyme catalyzes the reaction of methanol into the very toxic formaldehyde. The reaction mechanism is seen in Figure 4.

This subproject aimed to express the methane oxidation pathway in an Escherichia coli strain, to mimic natural methanotrophy of organisms such as M. capsulatus, This, in turn, could allow us to make use of the ribulose monophosphate (RuMP) pathway for formaldehyde assimilation [4]. As discussed in Chen et al 2021 [4], overexpressing three enzymes (methanol dehydrogenase, hexulose-6-phosphate synthase and 6-phospho-3-hexuloisomerase), can convert E. coli strains into methylotrophs that use methanol as sole carbon source. This new E. coli strain, named SM1 and expressing the heterologous RuMP pathway, is able to assimilate methanol into biomass, and as such is a perfect candidate for the biofilter. E. coli SM1 has a doubling time ranging from 8 to 55 hours, but can also grow on Lysogeny Broth (LB) medium, where the doubling time more closely resembles that of wild type (WT) E. coli.
Additionally, in this project another strain is used to mimic methanotrophy in E. coli. The C1-glycine-serine (C1GS) auxotrophic strain, here referred to as E. coli C1GSaux, , is not capable of growing in media without glycine or serine supplementation [5, 6]. This C1 strain can heterologously express the reductive glycine pathway (rGlyP). Through this linear C1GS-assimilation pathway, this strain is able to assimilate formate, and potentially methanol or methane, into biomass. As such, in absence of glycine, the C1GSaux strain requires formate, methanol or methane to assimilate glycine, serine and C1 biomass precursors from these one-carbon substrates .
C1 strains
- E. coli CFC526.0 ΔgapA::gapC ΔpfkA pFC139C
- E. coli SIJ488 C1(G)S-auxotroph (Δgcv ΔserA ΔfdhE) with genomic insertion of ftl-mtdA-fch + recovered frmAB (ΔyaiL)
Methanol growing strain with RuMP pathway (SM1) from F. Chen et al 2021 [4]
C1 auxotrophic strain (C1Saux) based on Yishai et al 2017. Genome integration of the ftl, mtdA and fch genes was performed recently by Bar-Even lab and is not published yet, but strain was kindly provided to us [5],
Approach
To tackle this pillar of the project, a multifaceted approach was used. We aimed to clone both pMMO and sMMO in an E. coli cloning strain, and finally in both the methanol growing strain (SM1) as well as in the C1 auxotrophic strain (C1Saux).
Particulate methane monooxygenase
In this project, the method described in Kim et al 2019 [7] for heterlogous expression of pMMO is used with minor alterations. In vitro activity of pMMO wass shown using a scaffold and duroquinol as additional reductant . The authors expressed several pMMO constructs on a high copy number plasmid, under control of a T7 promoter with a lac operator. pMMO is usually a membrane-bound complex. To mimic the active site of native pMMO in a soluble form, the pMMO-B1 and –B2 substructures were linked to an apo form of human-heavy chain ferritin (HuHF), which work as a scaffold. This was performed either in series or in parallel, resulting in the constructions of mimics pMMO-m1 to m4. The authors then measured methanol production, which works as a scaffold. In this project, pMMO-m1 was constructed again and tested in vivo and vitro.

The plasmid shown in Figure 5 was constructed through Gibson assembly. For this end, two G-blocks are ordered from IDT DNA, the first one carrying both huHF, His-tagged, and pmm0-B1, and the second harboring pmmo-B2. Additionally an empty pET-28a(+) plasmid is used from the Wageningen collection. This is transformed into a E. coli BL21 strain, which is specialized for protein expression (NEB).
After successful transformation of the resulting plasmid, the pMMO protein complex was purified through the His-tag attached to huHF. Activity of pMMO was then tested in vitro by running enzymatic assays. The enzymatic assays contained the reductant duroquinol, that was obtained following the reducing protocol as shown in Kim et al 2019 [7]. The described method, however, did not provide enough information to completely reproduce their approach. Thus, estimations were made regarding precise compound quantities. The assay also contained the pMMO protein complex in a Tris-buffered solution. As substrate, ¼ atmosphere of methane was held in an anaerobic bottle, together with ¾ air. After 25 hours, the methanol concentration was measured using GC. Bottles without duroquinol, enzyme or methane were used as negative controls. The protein was also tested in vivo by growing the E. coli BL21 strain harboring the pMMO-m1 plasmid in similar conditions to the enzyme assay on M9-glucose and measuring the methanol concentration every 24 hours.
Soluble methane monooxygenase
In this project, the method described in Bennet et al 2021 [8] was used to express sMMO in an E. coli strain, with slight alterations. In this paper, the authors took the entire sMMO operon from M. capsulatus (Bath) and transfered it into a medium-copy number plasmid under control of an araBAD inducible promoter from the SEVA (Standard European Vector Architecture) collection . In addition, the genes for chaperones GroEL-2 and GroES were taken from E. coli MG1655 and GroL and GroS from M. capsulatus (Bath), to ensure correct folding of the enzyme. These chaperones were placed under control of constitutive promoter J231XX, similar to the Anderson collection. Added to the registry as BBa_K3747004.
This approach is similar to the one of 2014 iGEM team Braunschweig’s, that also used the entire sMMO operon, but under control of the lacI-lambda pL hybrid promoter. (BBa_R0011). This team also expressed chaperone proteins from E. coli, similar to the 2021 article. However,in the work of Bennet et al., as well as in our approach, some chaperone proteins from M. capsulatus were also expressed. Although a similar approach to this article is taken, in this project some alterations were made, mainly regarding the plasmid backbones. Instead of the origin of replication (ORI) used in the original patent [9] and in the related article, more conventional medium copy-number ORIs from the SEVA collection plasmid backbones are used (Figure 6 and 7). The benefit of such backbones is their interoperability and experience within the Wageningen campus. Additionally, as can be seen in Figure 7, the gene coding for Mdh from Bacillus subtillis, from pZASSC-rbsC_bsMDH [10], was also incorporated in the chaperones plasmid. This limits the number of plasmids required for the E. coli C1Saux strain to become methanotrophic from 3 to only 2.


To construct the plasmids shown in Figure 5 and 6, Gibson assembly was used. First interim plasmids are constructed as shown in Table 1. These were assembled as parts to then construct the final plasmids. The sub-parts are obtained both from PCR reactions from E. coli MG1655 or GBlocks kindly provided by our sponsors IDT-DNA and Twist biosciences.

Growth experiments
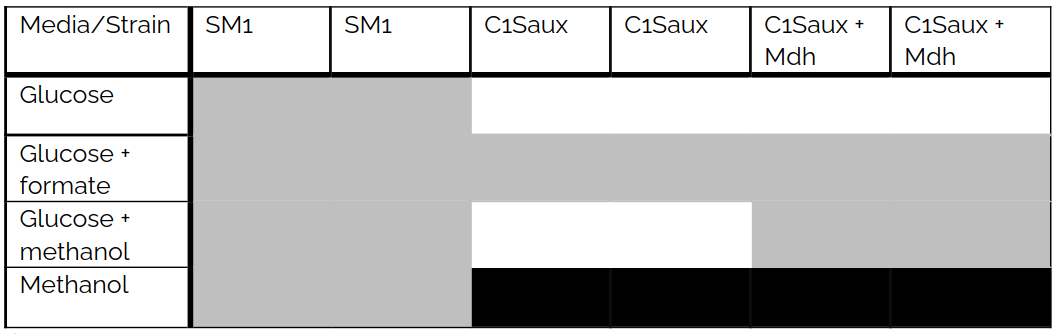
As mentioned before, to complete synthetic methanotrophy in E. coli the conversion of methanol into biomass and carbon dioxide is required. There are two strains in this project that are able to do some aspect of this process. The C1Saux strain can convert formaldehyde into biomass and is auxotrophic for formaldehyde (or formate) when grown on minimal media (M9). The second strain, the SM1 strain, can grow on MOPS minimal media and methanol as sole carbon source. Both growth requirements were verified in-house.
In this project the C1Saux strain was transformed with pZASSC-rbsC_bsMDH plasmid harboring Mdh to complete the conversion of methanol into biomass and carbon dioxide. To test this qualitative growth, experiments were run on minimal media with methanol or glucose as carbon sources, either alone or in combination.
Results
pMMO

After purification and SDS-PAGE electrophoresis, pMMO-m1 was confirmed as expressed (Figure 8). After this initial evidence, the purified enzyme was used in an enzymatic assay. After 25 hours of incubation with methane, no increase in methanol concentration was observed. In the in vivo test, an E. coli expressing the pMMO-m1 construct did not yield any increase in methanol concentration when grown for 24, 48, 72 or 96 hours in presence of methane. As such, no activity of this enzyme was reported.
sMMO
The interim plasmids pSEVA391_chap(1-2), pSEVA2610_sMMO(1-2), pSEVA2610_sMMO(3-4) were obtained in the E, coli cloning strain DH5α. However, due to time constraints, no final plasmid could be obtained.
Results growth experiments
The growth experiments showed the successful conversion of methanol into formaldehyde in the E. coli CGS1aux strain. When grown with glucose and methanol on M9, this strain grew as well as on glucose with formate on M9. On the other hand, growth was not observed on M9 with only glucose alone, indicating auxotrophy for C1 and presence of active Mdh. Additionally, when E. coli SM1 was grown on MOPS medium with 400 mM methanol as sole carbon source. These results are shown in Table 2.
Conclusion
The conversion of methane into methanol remains a bottleneck in synthetic methanotrophy. The soluble form showed problems in the cloning stage. However, as shown in the iGEM 2014 Braunschweig’s project, Clarke et al 2021 [9] and Bennet et al 2021 [8], expression is possible in vivo . The particulate MMO mimic 1 form showed problems both in vivo and in vitro , showing no enzymatic activity. It still remeains to be asserted whether this was due to the reductant duroquinol not being sufficiently reduced or due to our pMMO-m1 construct not being active. For now, we have shown the dependence of E. coli C1GSaux on C1-substrates and the successful expression of Mdh in this organism, allowing it to grow on methanol. The latter methanol-dependent strain provides a perfect selection platform to proof in the future the conversion of methane into methanol, either by sMMO or an active version of the pMMO mimic.
-
References
arrow_downward- Nguyen, Anh Duc et al. Trends in Biotechnology, Volume 39, Issue 4, 381 – 396 https://doi.org/10.1016/j.tibtech.2020.07.007
- Walters, K.J., Gassner, G.T., Lippard, S.J., & Wagner, G. (1999). Structure of the soluble methane monooxygenase regulatory protein B. Proceedings of the National Academy of Sciences of the United States of America, 96 14, 7877-82 .
- A.K. Shiemke, S.A. Cook, T. Miley, P. Singleton, Detergent Solubilization of Membrane-Bound Methane Monooxygenase Requires Plastoquinol Analogs as Electron Donors, Archives of Biochemistry and Biophysics, Volume 321, Issue 2, 1995, Pages 421-428, ISSN 0003-9861, https://doi.org/10.1006/abbi.1995.1413.
- Yan-Yu Chen, Yuki Soma, Masahito Ishikawa, Masatomo Takahashi, Yoshihiro Izumi, Takeshi Bamba, Katsutoshi Hori, Metabolic alteration of Methylococcus capsulatus str. Bath during a microbial gas-phase reaction, Bioresource Technology, Volume 330, 2021, 125002, ISSN 0960-8524, https://doi.org/10.1016/
- Oren Yishai, Leander Goldbach, Hezi Tenenboim, Steffen N. Lindner, and Arren Bar-Even ACS Synthetic Biology 2017 6 (9), 1722-1731 DOI: 10.1021/acssynbio.7b00086
- Kim, S., Lindner, S.N., Aslan, S. et al. Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat Chem Biol 16, 538–545 (2020). https://doi.org/10.1038/s41589-020-0473-5
- Kim, H.J., Huh, J., Kwon, Y.W. et al. Biological conversion of methane to methanol through genetic reassembly of native catalytic domains. Nat Catal 2, 342–353 (2019). https://doi.org/10.1038/s41929-019-0255-1
- R. Kyle Bennett, Nyaradzo Dzvova, Michael Dillon, Stephanie Jones, Kelley Hestmark, Baolong Zhu, Noah Helman, Derek Greenfield, Elizabeth Clarke, Eleftherios T. Papoutsakis doi: https://doi.org/10.1101/2021.08.05.455234
- Clarke et al (patent) 10,894,951 B2
- Wenk, S, Schann, K, He, H, et al. An “energy-auxotroph” Escherichia coli provides an in vivo platform for assessing NADH regeneration systems. Biotechnology and Bioengineering. 2020; 117: 3422– 3434. https://doi.org/10.1002/bit.27490

