




- design
- Biosynthesis of Cdh and CkTcS
- Cdh and CkTcS
- Global Transcription Machinery Engineering
- Temperature-sensitive suicide switch
- Plasmid Construction and SUMO tag
- Biosynthesis of SAM
- Directed Evolution
- Evaluating and Choice
- Cdh: Error-prone PCR
- CkTcS: PROSS Calculation
- Enzyme immobilization
- References
In wetlab, the three main goals were:
1. Obtaining all two enzymes and two coenzymes to construct a cell-free enzyme-catalyzed degradation system
2. Improving biosynthetic efficiency of enzymes and coenzymes
3. Verifying that this cell-free system can work effectively in a coffee environment
From our design, we successfully achieved biosynthesis of Cdh, CkTcS and CoQ(0) at higher efficiency, and which can be used in industrial production. At the same time, we use multiple ways to improve adaptation to the coffee environment of the cell-free system. You can find a detailed description of the design of our project below.
Biosynthesis of Cdh and CkTcS
Cdh and CkTcS
In the first step of the reaction, Cdh changes the 8’C of caffeine to carbonyl with the participation of coenzyme Q0.

Green dots represent nitrogen atoms and blue dots represent oxygen atoms
Cdh is caffeine dehydrogenase, composed of CdhA, CdhB and CdhC. We had obtained its complete gene sequence. However, the three-dimensional structure of Cdh is unclear. The Km of Cdh is around 4.5μM.
in the second step of the reaction, the N-methylation of the reciprocal isomer 1,3,7-trimethyluronic acid of the product of the previous step in the presence of CkTcS produces theacrine.

Green dots and brown represent nitrogen atoms and blue dots represent oxygen atoms
CkTcS consists of eight chains. We had obtained its complete gene sequence and the three-dimensional structure is shown below.

The caffeine concentration in coffee and other caffeinated beverages in general is much larger than the Km, and it can be assumed that our enzymes are able to catalyse reactions at the maximum reaction rate. The optimum pH for both enzymes are both 7.0, the optimum temperature for Cdh is around 66°C and the optimum temperature for CkTcS is 27°C. In order for our system to work properly in hot and acidic coffee solution we need to make directed evolutionary modifications to both enzymes.
Global Transcription Machinery Engineering
CRP (cAMP receptor protein), a protein with extensive transcriptional activation in Escherichia coli, functions at the initiation of transcription. The transcriptional efficiency of this gene can be altered by changing the ease of CRP binding to the upstream elements of the controlled gene. The target products of this project are two proteins, and the global transcriptional regulation part improves the expression efficiency of numerous genes in the bacterium by expressing exogenous CRP, thus improving the resistance and metabolic ability of the engineered bacteria. Advances in the study of using global transcriptional regulation engineering to improve strain tolerance and indirectly improve protein yield at mass production. To ensure transcriptional efficiency, both the promoter and 5'UTR sequence of the gene were replaced with the type with high transcriptional and translation efficiency mentioned in the literature.

click here to download plasmid of Global Transcription Machinery Engineering PUTRssrA-PUTRinfC-rplT.txt

Temperature-sensitive suicide switch
Although following the planned process, our engineered bacteria will not cause safety problems, we have added an insurance policy - a temperature-sensitive suicide switch - to prevent possible accidents and leaks in the lab.
We designed a temperature-sensitive suicide switch using toxin RelB and antitoxin RelE, and added it to the upstream of the two enzyme’s genes . In a scene of industrial production, as long as the temperature is down to below 35 degrees Celsius, the engineered bacteria will suicide immediately to avoid microbial leakage and contamination.

please click here to download our plasmid of Ptrc toxin-antitoxin safety module.

In the meantime, we would like to follow another temperature-sensitive suicide switch that UCAS-China involved in last year's project. In the gene circuit, CI434ts as a transcription factor can repress TEVts and doc toxin expression, while TEVts as a protease can cut CI434ts, thus forming a balanced relationship. And when the temperature decreases, CI434ts activity decreases and loses its inhibitory effect on downstream toxins, thus initiating the engineered bacteria suicide program.

For more information on this part, see our project last year.
Plasmid Construction and SUMO tag
Although we have added temperature control switch switch to our engineered bacteria to ensure biosafety, cell-free system is still the best option when considering coffee flavor and maintenance of our portable decaffi coffee cups. To achieve this goal, we need to introduce a useful molecular tag when designing the plasmid to enable efficient purification of the two target proteins.
We used SUMO tag, which was proved can improve the stability and solubility of recombinant proteins and reducing inclusion body production, thus improving the production efficiency of engineered bacteria. In addition, after the target protein is extracted from the cell fragmentation fluid, the SUMO tag can be cut off completely with the specific protease ULP1, getting a product that is identical to the original protein sequence.
In our plasmid, we attached SUMO tag to the N-terminal end of the target protein. To know more about our plasmid construction, visit Parts page.
You can also click the button below to download our Plasmid sequences. Cdh CkTcS

As shown in the figures below, the purification process is divided into three steps. In the first step, the fusion protein of SUMO and the target protein is bound to the Ni column with the help of the 6-his tag on SUMO and thus separated from the cell fragmentation solution; in the second step, after one day of enzymatic cleavage, the SUMO tag is cleaved off the target protein by the ULP1 enzyme; in the third step, the enzymatically cleaved mixture containing the three components (SUMO, ULP1 and the target protein) is again flowed through the Ni column again. Since both SUMO and ULP1 enzymes contain 6-his tags, they will both be bound to the column, so we only need to look for the target protein in the solution before eluting the protein from the column.



Biosynthesis of SAM
S-Adenosyl methionine(SAM) is a coenzyme in the methyl transfer reaction. Unlike enzymes, coenzymes are consumable and not reusable. Therefore, despite the fact that SAM is a small molecule, we are still eager to achieve efficient biosynthesis of SAM.

Green dots represent nitrogen atoms, blue dots represent oxygen atoms
and yellow dots represent sulphur positive ions
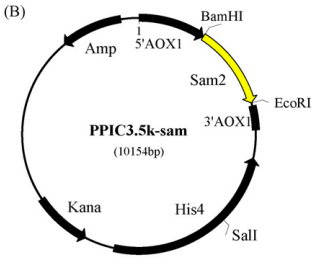
Yeast synthesis of SAM is mainly dependent on the dimerization and tetramerization of two enzymes, sam1 and sam2, of which the expression of sam2 is not inhibited by the product, so it was chosen to increase the synthesis of SAM by enhancing the expression of sam2 gene. This was done by first extracting the sam2 gene from Saccharomyces cerevisiae, double digesting it with EcoR1 and BamH1 and then treating it with plasmid pPIC3.5k using DNA ligase to obtain the pPIC3.5k-sam plasmid.
The conversion of SAM to cysteine can be catalyzed by cystathione β-synthase (CBS), a process that depletes SAM and decreases its accumulation. It was therefore decided to inactivate the CBS gene by inserting an antibiotic gene into it, thereby achieving a knockout of the CBS gene. The wild GS115 strain was first cultured, after which the CBS gene was extracted from the GS115 strain by PCR and the CBS gene was integrated into the pMD19T plasmid to obtain the T-cbs plasmid. The Zeocin gene and the T-cbs gene were then treated with Spe1 restriction enzyme, followed by ligase treatment to insert the Zeocin gene into the CBS gene to obtain the T-c-z plasmid containing the inactivated CBS gene.
The engineering bacterium chosen was the Pasteur Bilbao GS115 strain, due to its many excellent characteristics. SAM can be synthesised with the added methionine under conditions where methanol is used as an inducer.
please click here to download the sequence of CBS.
CBS inserted by Bleomycin.txt


Directed Evolution
At this point, our genetic circuit is already complete and the cell-free system is already constructed. However, if Cdh is incubated at 70°C for 30 minutes, the enzyme activity decreases sharply to 11% of the initial value, and CkTcS is in a similar situation, which indicates that we cannot decaffeinate coffee at high temperature. Coffee lovers are well aware of the impact of temperature on sensory properties of coffee. In order to expand the usage scenarios of our coffee cup and maintain the most of coffee flavor and user experience, we hope to make CkTcS and Cdh have better catalytic activity in high-temperature coffee. Therefore, we used two kinds of evolution to modify the optimum reaction temperature of the two enzymes.
Evaluating and Choice
To improve the thermal stability of both enzymes, two options were considered: firstly, targeted evolution of wild-type proteins using error-prone PCR techniques to obtain the desired target protein using high-throughput screening, but this approach requires features that facilitate screening, such as a rate of decrease in reactant concentration that is positively correlated with their thermal stability. The second is the use of PROSS software to predict mutation sites on proteins capable of improving thermal stability to obtain new protein sequences, but this approach requires the crystal structure of the protein to be known.
Cdh--Error-prone PCR
Cdh is missing necessary information in the PBD library so that it cannot be rationally designed by software. Therefore, error-prone PCR were used for directed evolution. The Taq enzyme used by PCR has a low fidelity, whereas the presence of Mn2+ in solution can further reduce this value. DNTPs competition exists in the process of base pairing, and most of the mutations are also derived from this competitive base mismatch. Increasing the concentration of Taq enzyme, prolonging the elongation time and increasing the concentration of Mg2+ can stabilize the mismatch and improve the mutation rate. The G-T swing base pairs formed between dGTP and T lead to mutation bias, which is undesired for random mutation induction. Decreasing the concentration of dATP and dGTP or increasing the concentration of dATP/dGTP can reduce this bias. Commercial Kits are available for error-prone PCR, but we attempted PCR of the target gene with a concentration gradient of 0-2mmol/L Mn2+ in solution, so as to realize the introduction of point mutations at different frequencies. After amplification, the error-prone PCR product will be linked into the pBlueScript plasmid and transformed into E. coli BL21 for culturing, inducing protein expression and purifying the Cdh enzyme with different mutations we want. The substrates of Cdh oxidase are caffeine and coenzyme Q0, and coQ0 before and after the reaction has obvious absorbance changes. According to this characteristic, we can react in a series of temperature gradients and screen out mutation types with better heat resistance.
CkTcS--PROSS Calculation
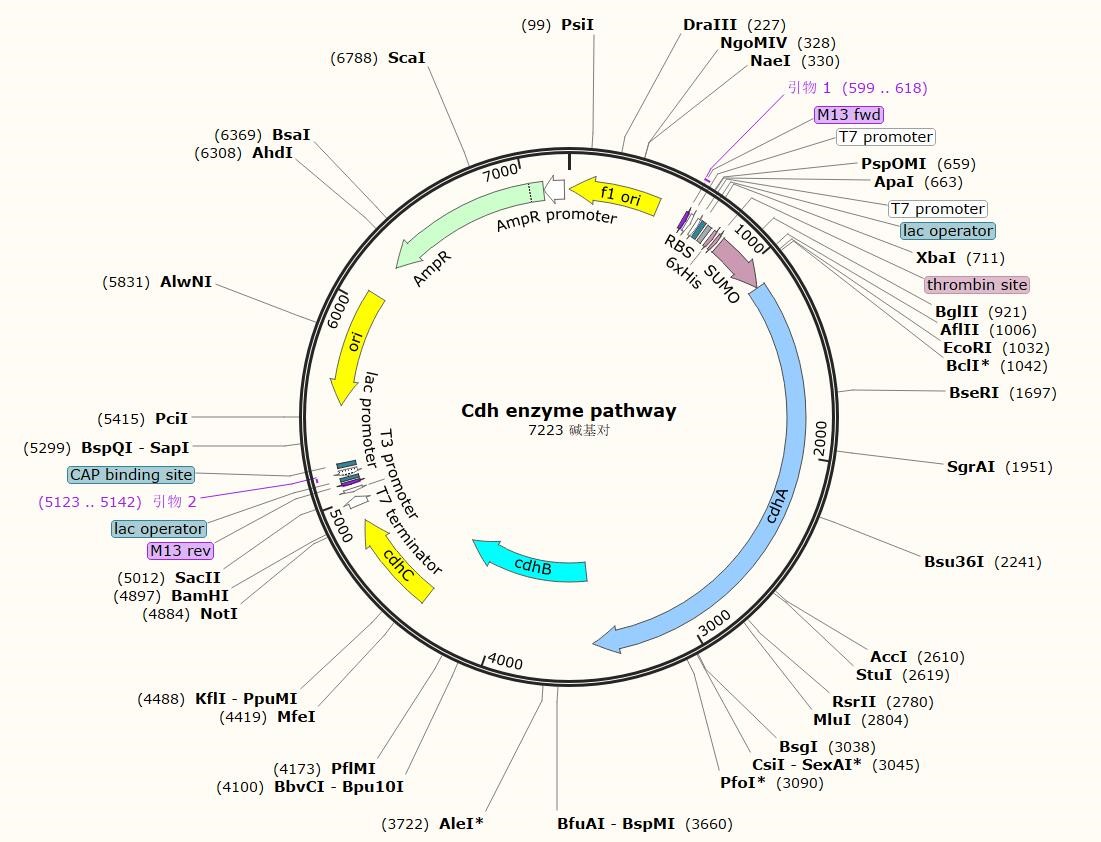
CkTcS has a known crystal configuration, which provides the opportunity for rational design using software PROSS. We used this platform to analyze the change of energy caused by the mutation sites, without decreasing its catalytic activity. According to the results, the plasmid was synthesized and verified in accordance with the same method of expressing the unmodified protein, confirming that we transformed CkTcS enzyme of better heat resistance without losing the catalytic activity. The specific design process will be detailed in the model section.

please click here to download the plasmid of our new CkTcS.
CkTcS enzyme pathway.txt

Enzyme immobilization
To use the enzyme CkTcs and Cdh efficiently and prevent them from leaking into the coffee, we need to immobilize these enzymes on the de-caffeine filter.
There are three common methods of enzyme immobilization.
1. Adsorption method
Adsorption is a method of immobilising enzymes or enzyme-containing organisms by adsorbing them onto their surfaces using various adsorbents. Commonly used adsorbents are activated carbon, alumina, diatomaceous earth, porous ceramics, porous glass, etc. The adsorption methodis simple, mild and does not cause denaturation or inactivation of the enzyme, and the carrier is cheap and easy to obtain and can be used repeatedly. However, the enzyme is not firmly bonded to the carrier and can be easily dislodged, so its use is limited.
2. Embedding method
Embedding is an immobilization method in which the enzyme is embedded in the pores of a polymer. This method usually uses polymeric substances such as polyacrylamide, starch, gelatin, etc. to form micro-mesh and thus immobilize the enzyme, or polymeric semi-permeable membranes to wrap the enzyme to form microcapsules. In this method, the enzyme is firmly bound to the carrier as in the adsorption method, but leakage may still occur and may also cause problems such as diffusion limitation.
3. Cross-linking method
The cross-linking method refers to the immobilization of enzymes by forming covalent bonds between enzyme molecules or between enzyme molecules and carrier molecules using some multifunctional cross-linking reagents, such as glutaraldehyde. As a chemical immobilization method, it can produce a stronger bond compared with physical methods, however, due to the low selectivity of the cross-linking agent and the high randomness of the connection, the construction of the active center of the enzyme may be affected, thus reducing the reaction rate or even directly inactivating the enzyme.
4. Covalent binding method
Covalent binding is an enzyme immobilization method that achieves irreversible binding by forming chemical covalent bonds between active functional groups on the surface of the carrier and non-essential groups on the enzyme molecule. Protein groups that can be coupled include amino groups, carboxyl groups, sulfhydryl groups of cysteine, hydroxyl groups of serine and threonine, etc. This is a strong binding method and although it may also affect enzyme activity, it causes less negative effects than cross-linking methods due to the ability to select non-essential groups of the enzyme molecule for attachment.
After comparing the advantages and disadvantages of these methods, we chose the covalent binding method. We used EDC and NHS to attach the enzyme to the carboxyl group of the acetate paper, which allowed us to minimize the possibility of its leakage with as little impact as possible on the enzyme activity. More importantly, it has been documented that the immobilized enzyme exhibits better heat and acid resistance in the coffee environment, which is of great for the process of caffeine removal.
References
Packer, Michael S.; Liu, David R. (2015). Methods for the directed evolution of proteins. Nature Reviews Genetics, 16(7), 379–394. doi:10.1038/nrg3927
Zhang, Y. H. et al. Identification and characterization of N9-methyltransferase involved in converting caffeine into non-stimulatory theacrine in tea. Nat Commun 11, 1473, doi:10.1038/s41467-020-15324-7 (2020).
Zeymer, C. & Hilvert, D. Directed Evolution of Protein Catalysts. Annu Rev Biochem 87, 131-157, doi:10.1146/annurev-biochem-062917-012034 (2018)
Guan, L. et al. Directed Evolution of Pseudomonas fluorescens Lipase Variants With Improved Thermostability Using Error-Prone PCR. Front Bioeng Biotechnol 8, 1034, doi:10.3389/fbioe.2020.01034 (2020).
Sammond, D. W. et al. An iterative computational design approach to increase the thermal endurance of a mesophilic enzyme. Biotechnol Biofuels 11, 189, doi:10.1186/s13068-018-1178-9 (2018).
Yang, M.-J., Lee, H. W. & Kim, H. Enhancement of thermostability of Bacillus subtilis endoglucanase by error-prone PCR and DNA shuffling. Applied Biological Chemistry 60, 73-78, doi:10.1007/s13765-017-0254-3 (2017).
Joo, J. C., Pack, S. P., Kim, Y. H. & Yoo, Y. J. Thermostabilization of Bacillus circulans xylanase: computational optimization of unstable residues based on thermal fluctuation analysis. J Biotechnol 151, 56-65, doi:10.1016/j.jbiotec.2010.10.002 (2011).
Cirino, P. C., Mayer, K. M., & Umeno, D. (2003). Generating mutant libraries using error-prone PCR. In Directed evolution library creation (pp. 3-9). Humana Press.
Yue-Hong Zhang, Yi-Fang Li, Yong-jin Wang et al. Identification and characterization of N9-methyltransferase involved in converting caffeine into non-stimulatory theacrine in tea. Nature Communications 2020,11:1473
Wang, L.-L., Fan, M., Xing, X., Liu, Y., & Sun, S. (2021). Immobilization of glyceraldehyde-3-phosphate dehydrogenase on Fe3O4 magnetic nanoparticles and its application in histamine removal. Colloids and Surfaces B: Biointerfaces, 205, 111917. doi:10.1016/j.colsurfb.2021.111917
Peroutka III R.J., Orcutt S.J., Strickler J.E., Butt T.R. (2011) SUMO Fusion Technology for Enhanced Protein Expression and Purification in Prokaryotes and Eukaryotes. In: Evans, Jr. T., Xu MQ. (eds) Heterologous Gene Expression in E.coli. Methods in Molecular Biology (Methods and Protocols), vol 705. Humana Press. https://doi.org/10.1007/978-1-61737-967-3_2
Bedade, D. K., Sutar, Y. B., & Singhal, R. S. (2019). Chitosan coated calcium alginate beads for covalent immobilization of acrylamidase: process parameters and removal of acrylamide from coffee. Food chemistry, 275, 95-104.





