Safety First
Overview
Safety has always been an important aspect of working in the laboratory. However, this year, safety became even more crucial due to the COVID-19 pandemic. Besides carrying out our protocols with the proper care for hazardous chemicals, following waste disposal procedures, handling biological materials appropriately and having always in mind our safety training, we paid special attention at all times to the sanitary measures imposed by government and institutional authorities.
The bacterial strains we were working with are not hazardous, not known to consistently cause disease in immunocompetent adult humans, and present minimal potential hazard to laboratory personnel and the environment. Nevertheless, we manipulated them to be genetically modified organisms (GMOs). So, if they escape the lab, they can be really hazardous due to the selection markers that we will use in our experiments. Horizontal transfer can occur within our strains and other microorganisms in the environment, which can lead to superresitant bacteria. That is why we were always really cautious to never let the strains escape the laboratory.
For more information, visit our completed Safety Form.

Lab Safety
We worked inside a Biosafety level 1 laboratory, with no risks for the environment and for humans, and with areas like open benches (Figure 2), chemical fume hood to manipulate hazardous chemicals (Figure 3), biosafety cabinet to ensure sterile conditions (Figure 1), and an eye washing apparatus in case accidents happened (Figure 4). We always followed the protective clothing rules: lab coats, gloves, safety glasses, closed shoes and long pants, as well as long hair tied back.
Also, we had a specific area for waste disposal (Figure 5) where we collected separately our biological, acid, alkaline, and acrylamide wastes, as well as sharp objects, for a qualified enterprise to retrieve them and discard them appropriately.
Biological waste was sterilized first before discarding.

Figure 1. Biosafety cabinet

Figure 2. Open benches.


Figure 3. Chemical fume hood.
Figure 4. Eyewashing apparatus

Figure 5. Waste disposal area
Sanity Measures
We wore a mask at all times and we tried to keep 1.5 m between each other while working at the open benches and/or inside the safety cabinets.
Only a few people were allowed to enter the lab because the institution had capacity restrictions, as shown in Figure 6.
Before entering and leaving the lab, everyone should wash or sterilize their hands. We had a table with sanitary products at the entrance (Figure 7).

Figure 6. Capacity restriction signal outside the lab

Figure 7. Sanitary products to sterilize our hands at the entrance and special trash can for masks
Our contribution to Safety in the Lab
We know that Safety is one of the most important aspects of working in a Lab. However, many of the protocols we make aren’t as safe as we think. For instance, the use of antibiotics is extremely controversial due to the impact it may have on generating antibiotic resistant bacteria. We wanted to address this problem through a satellite project using Synthetic Biology as a tool.
Just as we stated in the Project Description, our objective was to design a novel selection method for transformed bacteria. This would allow us to eliminate the use of antibiotics in genetic engineering protocols in the laboratory. Our designed marker includes the use of glycerol as a selection compound, a plasmid with a selection cassette with the gene katE and a modified strain of E. coli DH5⍺ which has the genes glpK, gldA and oxyR substituted by mRFP (as a reporter), Paralcaligenes ureilyticus Dehydratase (PuDHT) and alditol oxidase (aldO) genes. The modified strain can’t metabolize this compound without a selection cassette formed by a plasmid with a katE gene. In this new Metabolic Pathway, aldO will convert glycerol to peroxide and D-glyceric acid, katE will convert peroxide to water and oxygen and PuDHT will convert D-glyceric acid into pyruvate. Meanwhile, mRFP will act as a reporter. If the bacteria had a successful knockout, it would have a red coloration.
The design of the genes, the knockout protocol
For this section of the project, we designed a set of Biobricks to aid us in the knockout of E. coli DH5⍺. We used plasmid pSIJ8 for a Phage Recombination to aid us with this purpose (based on several papers that used this plasmid for their knockouts [1]). This plasmid has a set of proteins that if induced with arabinose, can begin the phage recombination with DNA fragments we insert. We decided to substitute the oxyR gene which activates the expression of hydrogen peroxide-inducible genes [2], with mRFP; gldA gene which is a key protein in the natural metabolism of glycerol, with PuDHT; and glpK which is also a key protein in the natural metabolism of glycerol, with aldO. The designs led to the creation of the parts shown here.
However, our parts didn’t arrive until after we entered the lab so, in order to get information about the concentration of glycerol at which we could inhibit proliferation, we did some experiments that involved the performance of an antibiogram and glycerol gradients.

Antibiogram - like assay
An antibiogram assay was performed with glycerol gradients ranging from 0 to 10% v/v from a separate water solution. Eleven assays were made, with increments of 1% starting from 0% of glycerol, serving as the positive control. The growth of DH5⍺ was analyzed in Petri dishes with LB divided into quadrants. Each quadrant contained a paper disc previously submerged in the glycerol solution of its respective concentration. As it can be observed, and as expected, the diameter of the halo of inhibition around the submerged placed paper disc grows as the glycerol concentration of the corresponding solution increases. This experiment would serve in future references with the aim to understand better and know the optimal concentration of glycerol at which DH5⍺ grows minimally, as well as to observe how it grows in other concentrations.
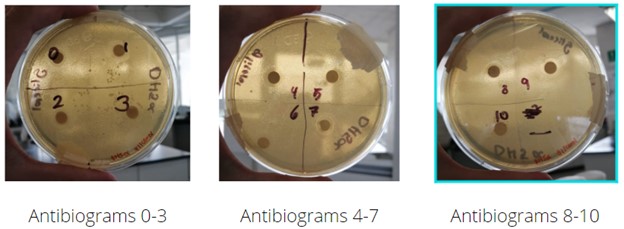
Figure 8. Results of the antibiogram-like assay
Normal growth can be observed in the control Petri dish quadrant with 0%v/v glycerol, this will serve as a guide to measure future growth in greater glycerol concentrations. Through concentrations 0 to 5%, the diameter of the halo presents similar length with only a slight increase in its measurement, meaning that DH5⍺ is able to grow without any problems at this glycerol concentrations. Beginning with a glycerol concentration of 6%, a more drastic increment in the length of the diameter of the halo can be observed, compared to the lower concentration assays. With 7, 8 and 9%, a rapidly increasing diameter is seen, while on 10%, no growth is observed near the paper disc.
Glycerol gradient pictures
Analysis of growth of DH5⍺, from cultured Petri dishes was performed. The experiment was carried out in duplicate, with LB as culture medium and glycerol concentration ranging from 0 to 20% (%v/v) in increments of 5%. As it can be observed, and as expected, the quantity of bacterial population diminishes as the glycerol concentration increases. The important factor in this experiment is to determine in which range of glycerol concentration %v/v does this bacterial strain show minimal growth, but still retains its cellular activity.


Normal growth can be observed in the positive control Petri dishes with 0%v/v glycerol, this will serve as a guide to measure future growth in greater glycerol concentrations. With 5%v/v a similar growth to the positive control Petri dishes can be observed in terms of density, but smaller populations are present. DH5⍺ shows almost none to null growth in the Petri dishes with 10%v/v glycerol, with clear null growth in higher glycerol concentrations. To be able to determine a more precise glycerol concentration in which DH5⍺ shows minimal growth with some difficulties, its recommended to perform this same assay with concentrations of glycerol ranging from 5 to 10%.
Glycerol gradient assay
As observed in the “Culture optic density through time at different glycerol concentrations” Graph, microbial growth is inhibited as glycerol concentration in the culture increases.
The culture medium where the glycerol concentration was 0% (LB) presented the highest optical density after 12 hours. Even 5 % of glycerol concentration on the culture medium decreased significantly the optical density after 12 hours. The curve of the culture medium with 20% glycerol concentration is practically flat, which demonstrates that glycerol at this concentration is the best to inhibit microbial growth.

Figure 9. Culture optic density through time at different glycerol concentrations.
The next graph shows how, as glycerol concentration increases, the part of the curve that corresponds to the exponential growth phase changes its behaviour. The slope decreases as glycerol concentration increases, which means that the difference between the optical density at the beginning and at the end of the growth exponential phase decreases as glycerol concentration increases, which proves a clear glycerol microbial inhibition effect. The highest optical density a culture may reach will keep decreasing as the glycerol concentration decreases.

Figure 9. Exponential phase of growth at different glycerol concentrations.
From the next graph, we can state that our experiments, measurements and conclusions are trustworthy, as the error in each glycerol concentration is small. The highest error value is shown in 20% glycerol concentration, but it is not greater than 0.15.

Figure 10. Growth error bars.
Genetic engineering - Knockout
PCR was carried out for our gBlocks puDHT, AldO, KatE, mRFP and pUC-ori. An agarose gel electrophoresis was performed to verify their sizes.

Figure 12. gBlocks digestions.
Then, gBlocks were ligated to the pSB1C3 backbone, and DH5a was transformed with the ligation products to be afterwards propagated in LB+CAM culture medium.
After that, PCR of mRFP, AldO and puDHT was performed and ran in an agarose gel electrophoresis in order to retrieve the DNA from the gel.
All fragments were the expected size, but the electrophoresis was repeated in order to obtain clearer bands.

Figure 14. PCR of gBlocks (second time).
Transformation of DH5a with eluted plasmids was carried out, and then PCR and agarose gel electrophoresis to verify that the gBlocks were really there.
All fragments were the expected size, so we concluded that the gBlocks are present in our DH5 cells.

Figure 11. gBlocks amplification.
Next, a digestion with restriction enzymes EcoRI and PstI, and HindIII and NcoI was carried out to make sure those enzymes cut the plasmids at the desired sites, producing expected fragment sizes.
In the last figure, we saw that all fragments were the expected size.

Figure 13. PCR of gBlocks (first time).
However, mRFP was the only visible band, so mRFP was the only DNA eluted from the second gel.

Figure 15. PCR of gBlocks after their transformation.
Finally, the katE gene was introduced to knockout DH5 cells. All of our constructions were inoculated in LB+7.5% glycerol culture medium. We had DH5 alone and transformed DH5 with pSIJ8 cells as controls. We also had DH5-KO and DH5-KO-plasmid (KO stands for knock out). Both cells with KO presented growth.
We were not expecting the cells with the KO and without the plasmid to grow because they didn’t have our selection cassette that contains catalase and allows bacteria to degrade hydrogen peroxide (product of the reaction of aldO) produced in response to glycerol. Nevertheless, we were able to prove that glycerol can be a good selection marker and we were able to synthesize genetic material that will give bacteria the ability to grow in a glycerol culture medium.
The implications of this can’t be underestimated, since we could expand the results we got here and use them to establish new protocols that don’t involve antibiotics. We will continue working on this project since we know that it is very important for us to improve Safety in the procedures we constantly make as students of Biology.
References:
[1] S. I. Jensen, R. M. Lennen, M. J. Herrgård, and A. T. Nielsen, “Seven gene deletions in seven days: Fast generation of Escherichia coli strains tolerant to acetate and osmotic stress,” Sci. Rep., vol. 5, no. 1, p. 17874, 2015.
[2] M. A. S. Toledo et al., “Characterization of an oxidative stress response regulator, homologous to Escherichia coli OxyR, from the phytopathogen Xylella fastidiosa,” Protein Expr. Purif., vol. 75, no. 2, pp. 204–210, 2011.
[3] S. Lindner et al., “A synthetic glycerol assimilation pathway demonstrates biochemical constraints of cellular metabolism”, The FEBS Journal, vol 287, 08 2019.


