DESIGN
Phase II--Optimization on cultural medium and thermal stimulus of Laccase CotA
Phase III--Improvement of Laccase CotA

In order to develop an enzyme-based cleaner, we determined enzymes utilized to degrade the main ingredients of gum base at first. In the first phase, we constructed basic parts BBa_K3990001, the coding sequence of Latex Clearing Protein from Streptomyces sp. K30, and Laccase CotA from Bacillus sp. HR03 targeting at degrading natural rubber and Ethylene-vinyl acetate copolymer (EVA) as representations of natural ingredients and synthetic ingredients of gum base, separately. Since the common gum base is currently made up of artificial ingredients, we decided to enhance the activity of Laccase CotA by optimizing the cultural medium and thermal stimulus, respectively, in the second phase. After that, two approaches, rational design and directed evolution, were applied to improve the substrate affinity and thermal stability of Laccase CotA, respectively. In conclusion, three engineering cycles, "Design → Build → Test → Learn → Design...", were established to accomplish the goal of degrading the gum base.
Phase I--Construction of LCP&Laccase CotA
Design at Level I
At this stage, we researched in order to solve the following problems:
1. What ingredients make the gum base nondegradable ?
2. What enzymes can be utilized to degrade the ingredients?
What ingredients make the gum base nondegradable?
As mentioned in Description, gum base, made up 15%-30% of gum, is the main non-digestible and water-insoluble ingredient of gum.[1] Although the detailed composition of gum base is the trade secret, the FDA approves 46 different chemicals used for manufacturing gum base.[2] These chemicals can be catalyzed into natural and synthetic materials as shown in the following table.

Table 1 The representative ingredients of gum base[4]
Latex Clearing Protein
Some gum bases are composed of natural rubber, of which chemical nature is polyisoprene. Researches were focused on enzymes that are capable of decomposing polyisoprene. These enzymes are displayed in Table 2. Both strengths and limitations were analyzed.

Table 2 Enzymes capable of decomposing polyisoprene
Considering the time limitation and the difficulty in obtaining specific strains expressing RoxA and RoxB, we chose LCP as the research object to decompose polyisoprene.
LCP oxidatively attacks the double bonds of polyisoprene molecules with the help of ferrous ion Fe2+ (Fig.1) to give cleavage products that have aldehyde and keto end groups as illustrated in Fig.2. The products of oxidized polyisoprene by LCP range from C20 tetra-isoprenoid to at least C35 hepta-isoprenoid as shown in Fig.2, which indicates that LCP works like endonuclease that can cleave the substrate at different positions.

Figure 1 | The mechanism of oxydizing polyisoprene by Lcp[1]

Figure 2 | The cleavage products of polyisoprene by LCP[2] and A UV(210-nm) absorbance spectrum of LCP products(gray line)[3]
Laccase CotA
Other gum bases are made of synthetic materials represented by EVA. We researched enzymes able to degrade EVA. The results are presented in Table 3.

Table 3 Enzymes capable of degrading polymer
From the table, it can be concluded that three types of enzymes all have good capabilities in degrading polymer. For alkB and Mn/Lignin peroxidase, help from co-expression and chemical oxidation play substantial roles in degrading polymer. In contrast, heterologously expressed laccase can degrade polymer to a large degree by itself, though the Laccase-mediator system increases the biodegradability.
Laccase, a common oxidase, is supposed to have broad substrate specificity. As EVA is a multi polymer, its accessibility of laccase active sites is limited. This limitation is overcome by the supplementation of mediator, ABTS (2, 2'-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) or 1-Hydroxybenzotriazole (HBT) as an electron carrier between laccase and substrate, as presented in Fig.4. The mediator is initially oxidized as an intermediate with high redox potential by laccase. Then, the mediator, a small chemical compound, gets access to oxidize the substrate more easily compared with enzyme. Therefore, the substrate EVA that cannot be oxidized directly by laccase is oxidized by the intermediate, while the oxidized mediator is reduced to its initial form. The mediator can maintain the cyclic redox conversion.[4] The overall oxidation system is called laccase-mediator system(LMS).
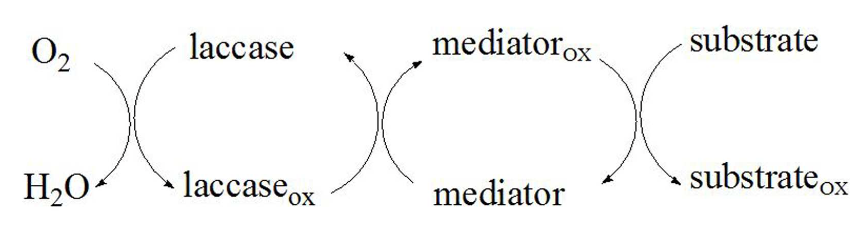
Figure 3 The laccase-mediator system
Phase II--Optimization on cultural medium and thermal stimulus of Laccase CotA
Design at Level II
As the main current ingredients of gum base are artificial, we decided to optimize the Laccase CotA, which is targeted at degrading EVA, a representative of synthetic ingredients. Literature reviews indicate that anion inhibition substantially influences the activity of Laccase CotA.[13] After reflecting on the methods of culturing, we planned to modify the cultural medium and supplemented metal ion. We changed Sodium Chloride (NaCl) in Luria–Bertani (LB) medium into Potassium Dihydrogen Phosphate(KH2PO4), and supplemented Copper(II) Chloride (CuCl2) into Copper(II) Sulfate(CuSO4). In addition to optimizing the cultural medium, we noticed that the thermal stimulus conditions of Laccase CotA can be tested as well. We wanted to set a inactivation curve related with two independent factors, temperature and time. According to the curve, the optimum thermal stimulus conditions of Laccase CotA can be determined, which would benefit the future study. In conclusion, we wanted to investigate the optimum cultural medium and thermal stimulus conditions of Laccase CotA.
In conclusion, we wanted to investigate the optimum cultural medium and thermal stimulus conditions of Laccase CotA.
Click here to learn more about experiments of Phase II.
Phase III--Improvement of Laccase CotA
Design at Level III
Based on the literature review, we utilized two approaches to improve the enzyme activity of Laccase CotA. One is rational design serving to modify the affinity between Laccase CotA, of which substrate is 2,2'-azino-bis(3-ethylbenzothiazoline-6-sul-fonic acid) (ABTS) and 1-Hydroxybenzotriazole (HBT), and the other one is directed evolution with the purpose of improving the thermal stability of Laccase CotA at ground temperature.
Rational Design
Computational protein design is a subset of the larger protein engineering field. Rather than rely on human intuition as has been done traditionally, computational protein design entails the modification and evaluation of an in silico protein model. The advantage of computational simulation is that it enables rapid testing and iteration of design tasks before slow and expensive laboratory experiments. [14]
With the help of modeling and experts, we identified the probable 3D-structure of Laccase CotA by homology modeling, the potential binding sites on Laccase CotA by molecular docking, and verified the binding sites by multiple sequence alignment. Then, recommended mutations on Laccase CotA, which can improve substrate affinity compared with original enzyme, were computed by molecular docking software. Among suggested single mutation, double mutations, and triple mutations, we chose three most stabilized ones to construct parts of mutated Laccase CotA. The measurement of constructed parts would be presented as well.
Click here to see expert interview of rational design. Click here to learn more about rational design.
Directed Evolution
Directed evolution aims to simulate the Darwinian evolution process in a test tube. Abundant and diverse mutants are artificially created through random mutations. These mutants are subsequently screened according to specific metrics with the goal of transforming proteins to obtain the specific functionality. Directed evolution is highly efficient as it enables the introduction of random mutations without understanding the underlying structure and function of the enzyme and the catalytic mechanism.[15]
Assisted by experts, we made a plan of conducting directed evolution experiments. Randomly mutated sequences coding Laccase CotA would be yielded from Error Prone-PCR(EP-PCR). Then, single colonies expressing mutated Laccase CotA would be cultured. The enzyme activity test on crude enzymes aimed at screening for beneficial variations. Next, those colonies obtained beneficial variations would be further cultured in shaken flasks. Finally, the enzyme activity results of purified mutated Laccase CotA would be compared with those of original enzymes.
Click here to see expert interview of directed evolution. Click here to learn more about directed evolution.
Reference
[1] Estruch, RA (2008). "Gum base". In Fritz, D (ed.). Formulation and Production of Chewing and Bubble Gum(2 ed.). Essex: Kennedy's Publications Ltd. pp. 93–118.
[2] e-CFR: Title 21: Food and Drugs PART 172—FOOD ADDITIVES PERMITTED FOR DIRECT ADDITION TO FOOD FOR HUMAN CONSUMPTION Subpart G—Gums, and Related Substances, retrieved 2016-12-09
[3] Xiuli Peng. Research on the removal of urban dirt. Beijing University of Technology,2004.
[4] "CFR – Code of Federal Regulations Title 21". www.accessdata.fda.gov. Retrieved 15 December 2016.
[5] Birke J, Jendrossek D. Rubber oxygenase and latex clearing protein cleave rubber to different products and use different cleavage mechanisms. Applied and Environmental Microbiology. 2014;80(16):5012-5020.
[6] Birke J, Röther W, Jendrossek D. RoxB Is a Novel Type of Rubber Oxygenase That Combines Properties of Rubber Oxygenase RoxA and Latex Clearing Protein (Lcp). Appl Environ Microbiol. 2017 Jun 30;83(14):e00721-17. doi: 10.1128/AEM.00721-17. PMID: 28500046; PMCID: PMC5494620.
[7] Naing SH, Parvez S, Pender-Cudlip M, Groves JT, Austin RN. Substrate specificity and reaction mechanism of purified alkane hydroxylase from the hydrocarbonoclastic bacterium Alcanivorax borkumensis (AbAlkB). J Inorg Biochem. 2013 Apr;121:46-52. doi: 10.1016/j.jinorgbio.2012.12.012. PMID: 23337786; PMCID: PMC3595352.
[8] Hyun Jeong Jeon, Mal Nam Kim. Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. International Biodeterioration & Biodegradation. Volume 103, 2015, Pages 141-146, ISSN 0964-8305. doi: 10.1016/j.ibiod.2015.04.024.
[9] Yoon, Moon & Jeon, Hyun. (2012). Biodegradation of Polyethylene by a Soil Bacterium and AlkB Cloned Recombinant Cell. Journal of Bioremediation and Biodegradation. 03. doi: 10.4172/2155-6199.1000145.
[10] Mukherjee, Shritama & Kundu, Patit. (2014). Alkaline Fungal Degradation of Oxidized Polyethylene in Black Liquor: Studies on the Effect of Lignin Peroxidases and Manganese Peroxidases. Journal of Applied Polymer Science. 131. doi: 10.1002/app.40738.
[11] Nishida, Tomoaki & Fujisawa, Mikihito & Hirai, Hirofumi. (2001). Degradation of Polyethylene and Nylon-66 by the Laccase-Mediator System. Journal of Polymers and the Environment. 9. doi: 10.1023/A:1020472426516.
[12] Santo, Miriam & Weitsman, Ronen & Sivan, Alex. (2013). The role of the copper-binding enzyme – laccase – in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. International Biodeterioration & Biodegradation. 84. 204-210. doi: 10.1016/j.ibiod.2012.03.001.
[13] Mohammadian M, Fathi-Roudsari M, Mollania N, Badoei-Dalfard A, Khajeh K. Enhanced expression of a recombinant bacterial laccase at low temperature and microaerobic conditions: purification and biochemical characterization. J Ind Microbiol Biotechnol. 2010 Aug;37(8):863-9. doi: 10.1007/s10295-010-0734-5. Epub 2010 May 15. PMID: 20473548.
[14]Adolf-Bryfogle, J., Teets, F. D., & Bahl, C. D. (2021). Toward complete rational control over protein structure and function through computational design. Current opinion in structural biology, 66, 170–177. https://doi.org/10.1016/j.sbi.2020.10.015
[15]Wei Xiong, Bo Liu, Yujiao Shen, Keju Jing, Thomas R. Savage, Protein engineering design from directed evolution to de novo synthesis, Biochemical Engineering Journal, Volume 174, 2021, 108096, ISSN 1369-703X, https://doi.org/10.1016/j.bej.2021.108096.



