
Trans-cleavage activity of AsCas12a
As part of a contribution to iGEM and future teams, we have added new information regarding the trans-cleavage activity of AsCas12a to a part BBa_K2965021 from ZJU-China iGEM 2019.
Based on a paper published by a research team in Nanyang Technological University (NTU), a test was conducted to examine the sensitivity of various Cas12 enzymes[1]. AsCas12a is able to detect for COVID19 RNA at the S2 site when the guide RNA perfectly matches the sequence. In the experiment, the gRNA sequence that was used is ACTCCTGGTGATTCTTCTT. Based on figure a, AsCas12a is able to specifically detect for COVID19 as there is no cross-reactivity for SARS-CoV or MERS-CoV. At 37°C, the fluorescence signal for SARS-CoV-2 is twice as high as compared to the fluorescence signal at 24°C.
In the paper, 10 different guide RNAs were made to test for mismatched activity of the different Cas enzyme. It was identified that AsCas12a produces minimal fluorescence reading when there is a mismatch of 1 nucleotide. This shows that AsCas12a is sensitive to a single nucleotide discrimination. This is also true for the LBCas12a.
Based on a paper published by a research team in Nanyang Technological University (NTU), a test was conducted to examine the sensitivity of various Cas12 enzymes[1]. AsCas12a is able to detect for COVID19 RNA at the S2 site when the guide RNA perfectly matches the sequence. In the experiment, the gRNA sequence that was used is ACTCCTGGTGATTCTTCTT. Based on figure a, AsCas12a is able to specifically detect for COVID19 as there is no cross-reactivity for SARS-CoV or MERS-CoV. At 37°C, the fluorescence signal for SARS-CoV-2 is twice as high as compared to the fluorescence signal at 24°C.
In the paper, 10 different guide RNAs were made to test for mismatched activity of the different Cas enzyme. It was identified that AsCas12a produces minimal fluorescence reading when there is a mismatch of 1 nucleotide. This shows that AsCas12a is sensitive to a single nucleotide discrimination. This is also true for the LBCas12a.
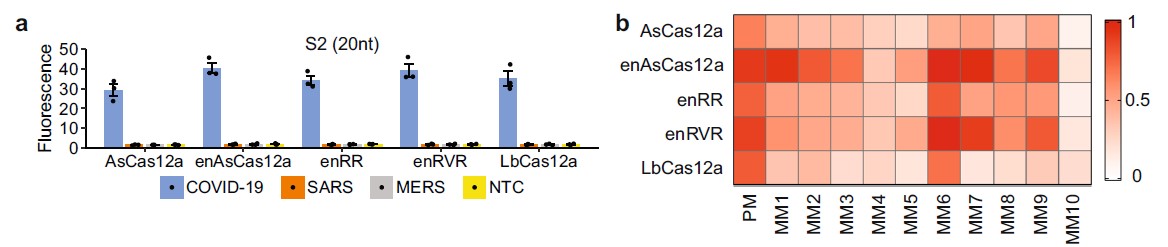
A. Intensity of the fluorescence produced for different Cas12a enzymes using single S2 gRNA sequence. B. Heatmap showing the intensity of the fluorescence produced after trans-cleavage assay was performed at 37 °C. 1 - highest fluorescence intensity measured & 0 - lowest fluorescence intensity
Reference
- K. H. Ooi, M. M. Liu, J. W. D. Tay, S. Y. Teo, P. Kaewsapsak, S. Jin, C. K. Lee, J. Hou, S. Maurer-Stroh, W. Lin, B. Yan, G. Yan, Y.-G. Gao, and M. H. Tan, “An engineered CRISPR-CAS12A variant and DNA-RNA hybrid guides enable robust and rapid covid-19 testing,” Nature News, 19-Mar-2021. [Online]. Available: https://www.nature.com/articles/s41467-021-21996-6. [Accessed: 21-Oct-2021].
