Results

Results
CloningThe initial step of the project was to clone our desired genes into 3 plasmids, i.e. plastic-degrading PETase/MHETase plasmid for E. coli, sucrose-uptake plasmid also for E. coli, and sucrose-secretion plasmid for S. elongatus.

Colony PCR of lacI and cscB: lanes 1 and 2 show a size of ~1.1 kb, while lanes 3-6 are ~1.2 kb, and lanes 7-10 ~1.5 kb. LacI is ~1150 bp, and with added flanked regions by primers, it should be ~1.3 kb. Lanes 3-6 show the correct lacI fragment. Lanes 7-10 show the correct cscB fragment.
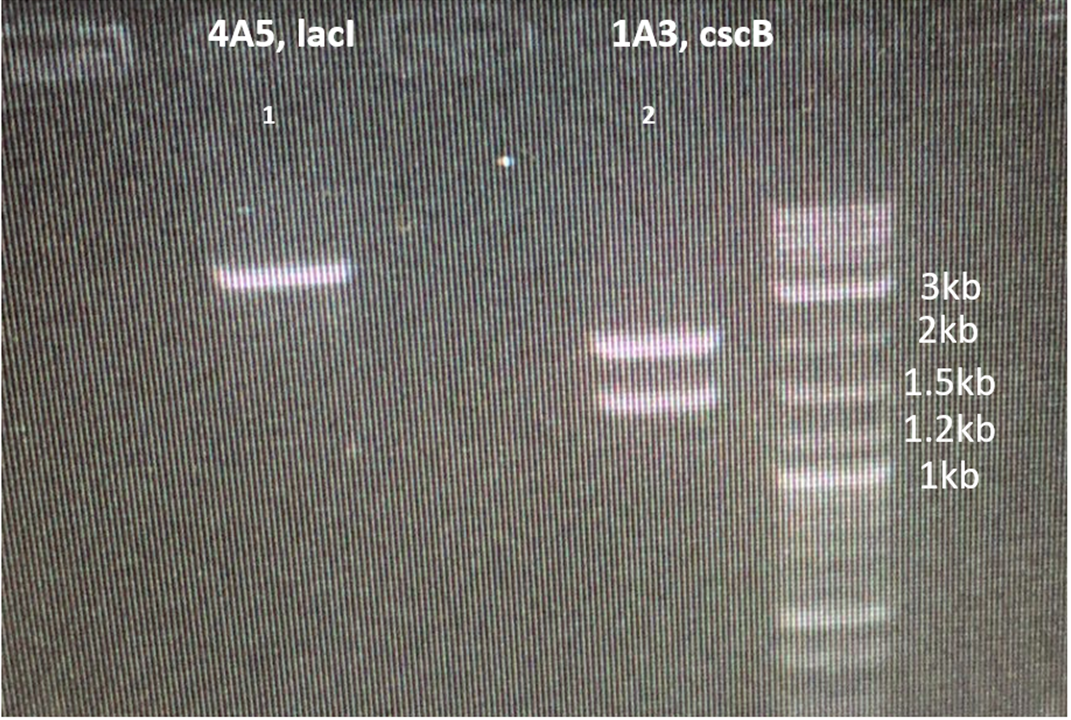
Restriction digestion analysis of lacI and cscB plasmids: Lane 1 with 4A5 and lacI plasmid and a single RE cut shows an expected ~4 kb (4.5 kb) band, while lane 2 shows cscB (1.5 kb) and 1A3 (2.1 kb). Lanes 1 and 2 both show the successful cloning of lacI and cscB gene fragments respectively.

Restriction digestion analysis: lanes 1-5 show RE digestion of cscA with ampicillin backbone, while lanes 6-10 show that of cscB. Lanes 3, 4 and 8 seem to show correct bands, but sizes of DNA were too close to be separated in gel. After running for additional time to separate them, lanes 3, 4 show the correct band size of 1.8 kb (cscA) and 2.1 kb (1A3 backbone), and lane 8 with 2.7 kb (cscB + K) and 2.1 kb (1A3 backbone). Lane 5 shows a RFP contamination.

Colony PCR and restriction digestion analysis: (Left) lanes 1, and 2 show colony PCR of cscA single fragment plasmid, while lanes 3 shows that of cscB+K. Lane 5 and 6 are cscA X/P digestion and cscB+K S/P digestion respectively. Lane 1 showed no bands, while lane 2 showed the expected size of 1.8 kb (cscA). Lane 3 showed an expected size of 2.7 kb. Lanes 5 and 6 both showed correct digestion bands, indicating successful clones. With a successful clone proven by lane 5, lane 1 is thus possibly PCR error. (Right) The experiment was repeated and positive results were observed for lanes 1-5, but S/P digestion band was larger than expected. Troubleshooting identified a problematic backbone.

Restriction digestion analysis of standard Biobrick assembly: lanes 1-8 show an E/P restriction digestion of a cscA and B+K plasmid. While the expected size should be 2 and 4.5 kb, all lanes except for lane 6 show bands of ~2.5 kb and ~4 kb. Lane 6 shows bands that are closest to the expected sizes, indicating a correct clone and thus a complete BL21 sucrose plasmid.
Cyanobacteria Growth
While PCC 7942 grows relatively slowly, we have acquired UTEX 2973 and plotted its growth in a 30°C illuminated shaking incubator. The optical density (O.D.) is measured daily, and its log(OD) value is plotted against time.


General Troubleshooting
While cloning is successful, our team encountered a number of caveats along the way, one of which is a constant and persistent RFP contamination. We tried our best to maintain lab hygiene and obtain clean backbones by repeating gel purification processes, but most of the time we were met with the surprising contamination of red cultures and colonies.

Moreover, we would often get yeast colonies on our bacterial plates. Yeasts generally grow slower than E. coli, but an easier differentiation is that yeasts normally appear more domed, white, and opaque than E. coli, as seen on the image on the right-most.

We speculated that the competent cells, incubator, or lab environment was/were contaminated with yeast spores. Working without a bunsen burner until August, transformation used to be done with a simple alcohol lamp. Moreover, with a lack of working fridges and freezers, we were required to transport of reagents from different labs, which increased chances of contamination. Plates were kept at a fridge, but it might not be cold enough to ensure the antibiotics' integrity on agar plates. These along with limited access to facilities such as Nanodrop, ice machine, a -80C fridge for competent cells), or stable access to shaking incubator (with light) led to our long troubleshooting and iteration cycles.
