Engineering

Rationale
Our project aims to develop a E. coli and S. elongatus co-culture system, where S. elongatus can photosynthetically metabolise sucrose, which fuels the production and secretion of PETase and MHETase in E. coli.A plasmid that codes for PETase and MHETase is transformed into E. coli. PETase digests PET into monomeric MHET and trace amounts of BHET, while MHETase further digests MHET into terephthalic acid and ethylene glycol. While a few E. coli strains can readily uptake sucrose as an energy source (eg. E. coli W), we have engineered the BL21 strain we are using to incorporate sucrose-utilising genes, i.e. sucrose permease, hydrolase, and fructokinase, into the bacterium genome through the lambda Red system. This ensures the function would not be easily lost through the generations as with exogenous plasmids. Sucrose produced by S. elongatus can be used by E. coli. We would also like to maximise sucrose production by S. elongatus through expressing cscB, which codes for sucrose permease that allows more sucrose transport in and out of the cell.
Our project is based on previous literature that has demonstrated successful co-culturing of E. coli and S. elongatus, and PETase/MHETase expression in E. coli [1, 2, 3, 4]. With the hardware and each function working as intended, the system should be able to maintain a long-term equilibrium between the two organisms, and produce enough PETase and MHETase to digest PET plastic.
Due to limitations in obtaining lab consumables and access to equipment, we were unfortunately unable to follow through with our experimental design. Our complete experimental design is detailed below.
Design
This project involves several components, which will first be tested and quantified individually, and then in a co-culture system.Plasmids
Three plasmids will be synthesised and constructed for our system:
1. Plastic plasmid: this plasmid contains coding regions for PETase and MHETase, two enzymes that will digest PET plastics into monomeric terephthalic acid and ethylene glycol.

2. Sucrose-uptake plasmid: this plasmid contains coding regions for the cscA, cscB and cscK genes, which are sucrose-6-phosphate hydrolase, sucrose permease, and fructokinase respectively. cscA allows for the metabolism of sucrose; cscB is a permease that allows sucrose entry into the cell; and cscK further aids in the uptake and digestion of sucrose. This plasmid allows E. coli to utilise sucrose as an energy source in addition to glucose.

3. Cyanobacteria plasmid: this plasmid contains coding regions for the lacI gene and cscB, a sucrose permease that can allow sucrose to pass through the cell membrane. The lacI gene creates an IPTG-induced system that stimulates the cyanobacteria to secrete sucrose when needed.

These plasmids are assembled using standard biobrick assembly methods.
- All parts synthesised by IDT and Twist Bioscience contain biobrick suffix and prefix regions, which can be recognised by specific restriction enzymes such as EcoRI, PstI, XbaI and SpeI.
- A standard digestion protocol is followed to create sticky ends for each insert and the corresponding backbone gene fragment. This is followed by a standard ligation protocol and chemical transformation of plasmids into DH5a (DE3) competent cells.
- Plasmids are confirmed by colony PCR or restriction digestion, and isolated and purified with a standard minipreparation kit/protocol.
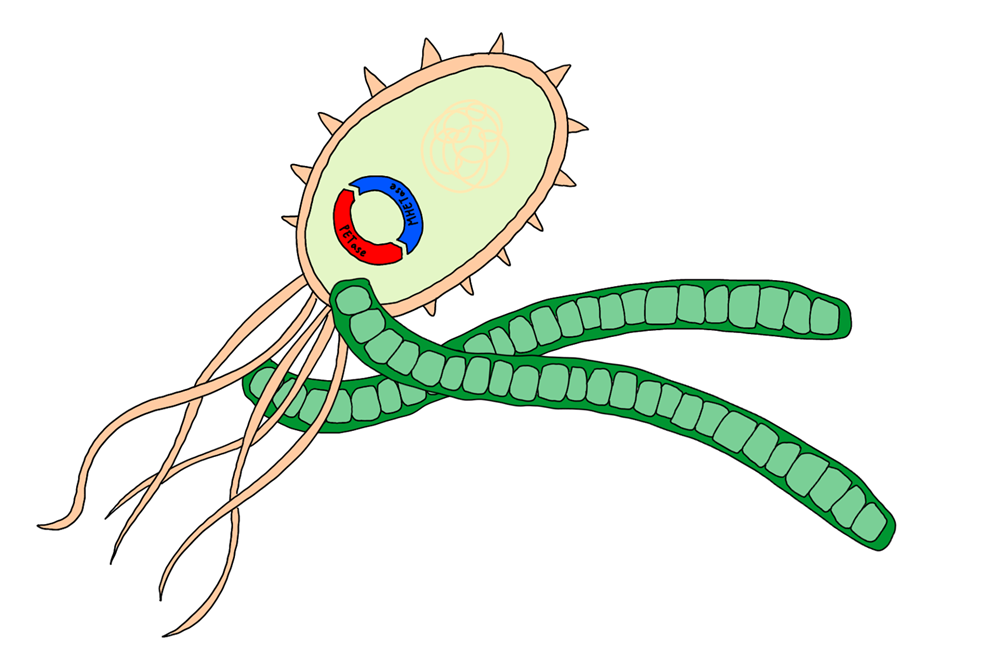
Co-culture of PET-degrading/sucrose-utilising E. coli and sucrose secreting cyanobacteria
Experimental design is separated into 4 main stages: 1) co-culture of E. coli and S. elongatus, 2) PET-degrading abilities of E. coli, 3) sucrose-utilising abilities of E. coli, and 4) sucrose secretion by S. elongatus.1. Co-culture
While direct of co-culture of E. coli and S. elongatus is possible and has been done by multiple research groups as well as iGEM teams, maintaining equilibrium requires a functional sucrose-utilising E. coli strain [5, 6, 7, 8] such as E. coli W, which we were unable to source. Furthermore, Hays et al. (2017) noted in their paper that wild type E. coli W grows most optimally when presented with more than 5g/L of sucrose, while cscB-enhanced S. elongatus produces merely 36 mg of sucrose per litre per hour. As a solution, the team had to further engineer E. coli W to adapt to a low-sucrose condition [9].At a very initial stage of our project, we decided that compartmentalising the two organisms would serve a better purpose for our prototype model. This allows the easy separation and exchange of nutrients and organisms.
To allow the exchange of nutrients, i.e. sucrose from S. elongatus to E. coli, we design a compartment with dialysis membranes between the organisms. Dialysis membranes have pores that are generally 1 to 10 nm in diameter, whereas sucrose molecules are 1.08631 nm in diameter. As a result, two compartments with E. coli and S. elongatus separated by a dialysis membrane would theoretically function well. From previous literature, such as one by Hays et al. (2017), cyanobacteria have been experimentally co-cultured with multiple organisms, including E. coli. Such co-cultures require an alternate BG-11 recipe that can support E. coli growth, which was named (CoB)BG-11 [10]. In other words, both compartments would contain the same medium of (CoB)BG-11.
In the same study that looked at S. elongatus and E. coli co-culture, the researchers noted that the long-term stability between these two organisms could be due to E. coli developing the ability to live off other metabolites produced by the cyanobacteria, and that it is not solely dependent on sucrose as a carbon source. Therefore, precaution should be taken against any extra nutrients introduced into the system that can alter the co-culture. This especially applies to the system’s implementation, since the adaptability of E. coli to utilise any immediate energy source could cause trouble if it leaks into open waters.

1.1 Cyanobacteria
Cyanobacteria works as the energy factory in our system by converting sunlight into sucrose for our bioengineered E. coli (BL21(DE3)). For our system, the species and strain of cyanobacteria used is S. elongatus (PCC 7942). To enable the construction of an accurate co-culture model, it is essential to know the growth conditions and growth rate of PCC 7942.
1.1.1 Growth conditions
PCC 7942 is a freshwater strain that can be cultured in BG-11 medium (freshwater). However, while liquid culture protocols are widely established, there are a lot of different solid medium recipes available online. Since solid cultures are more conveniently used in experiments and often utilised for preservation (of up to 2 months), we took different recipes and tested them with PCC 7942, i.e. different concentrations of agarose powder (1.5%, 1.25%, 1% and 0.75%), and using different types of agar like bacto agar. Colonies formed 2 weeks after streaking on 1.5% BG-11/agar gel. However, due to this slow growth rate of PCC 7942 and limited resources to accommodate our plates, we reverted to liquid cultures for further experiments.

PCC 7942 grows well under 30°C with bright light. We placed tubes of cyanobacteria in a shaking incubator (200 rpm) under a constant light source, and supplemented them with fresh BG-11 medium when cultures became opaque or too concentrated.
1.1.2 Growth rate: measurement of OD
To effectively quantify the number of cyanobacteria, we implemented microbial OD measurement. We measured the OD of cyanobacteria tubes every day at a set time.
An alternative method of measurement would be to use counting chamber slides and manually count the number of cyanobacteria cells. A serial dilution would then be employed to obtain the data necessary for generating a growth curve.
2. PET degradation
To test the function of PETase plasmid and the produced PET enzyme, a digestion experiment would be conducted. For PETase secretion, we directly check the degradation rate of PET plastic strips in culture medium, as performed by previous iGEM teams as well [11]. We also test the efficiency of the plasmid in different strains of E. coli, i.e. DH5a(DE3), a strain optimised for cloning, and BL21(DE3), a strain optimised for protein expression.After transforming the plasmid into each strain, we pick a colony and culture them in 5mL of LB medium in a 37°C shaking incubator. At the same time, we add 10 PET squares (2cm x 2cm) into the culture tube. Every 12-16 hours, we take out 1 square of PET and image the surface with scanning electron microscopy. We will also add 5mL of fresh LB medium into the culture during this time. After 20 days, all strips will be taken out for imaging. This will give us a time-dependent insight into PETase activity and provides us with a preliminary idea for the speed and efficiency of wild type PETase. Alternatively, we can take 1mL of the previous day’s culture and transfer it to a new tube along with all strips of PET, while adding 4mL of fresh LB medium. This ensures that E. coli does not overgrow.
This test will be adjusted based on experimental results. The aim of adding fresh LB medium is to ensure E. coli continues to grow and produce PETase at a steady rate. However, it is difficult to maintain bacteria and enzyme concentration at a constant. While it is possible to heat kill E. coli at 60°C for 5 minutes and leave PETase active (denaturing temperature at 69.4°C), the co-culture design does not allow for this method [12]. It is also advisable to extract PETase from E. coli and directly test with the enzyme instead of testing with the bacterial chassis as well. After extraction of PETase, as there is no need to switch out medium and plastic trips, we are able to check how long it takes for the enzyme to digest PET strips until there is a significant and measurable difference after imaging.
Should the resources be available, we can also perform high-performance liquid chromatography (HPLC) to check the presence of PET monomers in the supernatant of the enzyme-plastic mixture instead. This is a more direct way of making sure that PETase is functional, since it can detect trace amounts of terephthalic acid and ethylene glycol that might not be visible under a microscope.
3. Sucrose uptake of E. coli
We then test the sucrose uptake ability of E. coli by measuring its growth rate when presented with different concentrations of sucrose and glucose. After transforming E. coli with a sucrose plasmid (with csc A, csc B, and csc K genes that enables sucrose metabolism), we culture it in different concoctions, i.e. LB, LB with glucose, LB with sucrose, LB with both glucose and sucrose etc. We measure the OD of the solution after incubation overnight at 600 nm to determine how well E. coli can survive in each situation. The experimental procedures are adjusted according to results. With enough data, a growth curve is constructed to determine optimum sucrose concentration. This allows us to construct a co-culture ratio between E. coli and S. elongatus that can allow them to be sustainably maintained. According to literature, while S. elongatus is able to generate 36 mg sucrose L−1 hr−1, and E. coli takes in sucrose at 7mmolgCDW−1 hr−1, an equilibrium of 1:10 for E. coli and S. elongatus is possible [13, 14].
4. Sucrose secretion of cyanobacteria
To test whether S. elongatus secretes more sucrose after being engineered, we use a sucrose kit to compare the sucrose content at set time points. We should also note that the over-expression of cscB lowers S. elongatus growth [15].The literature we base our modelling off of uses a 48-hour constant light interval for cyanobacteria growth [16]. This can be tested against a 12-hour cycle, which better mimics the natural environment and would provide a better estimate of the model working at a real-life condition.
After we verify this, we circle back to Stage 1 and test the co-culture system with the now-enhanced sucrose secretion efficiency of S. elongatus. We can then establish an appropriate co-culture ratio, followed by verification of the PET degradation efficiency and application of this system in our proposed hardware prototype.
References
[1] Hays, S. G., Yan, L. L., Silver, P. A., & Ducat, D. C. (2017). Synthetic photosynthetic consortia define interactions leading to robustness and photoproduction. Journal of Biological Engineering, 11(1). https://doi.org/10.1186/s13036-017-0048-5.[2] Seo, H., Kim, S., Son, H. F., Sagong, H.-Y., Joo, S., & Kim, K.-J. (2019). Production of extracellular petase from Ideonella sakaiensis using SEC-dependent signal peptides in E. coli. Biochemical and Biophysical Research Communications, 508(1), 250–255. https://doi.org/10.1016/j.bbrc.2018.11.087.
[3] Shi, L., Liu, H., Gao, S., Weng, Y., & Zhu, L. (2021). Enhanced extracellular production of ispetase in Escherichia coli via engineering of the PELB signal peptide. Journal of Agricultural and Food Chemistry, 69(7), 2245–2252. https://doi.org/10.1021/acs.jafc.0c07469.
[4] Janatunaim, R. Z., & Fibriani, A. (2020). Construction and cloning of plastic-degrading recombinant enzymes (MHETase). Recent Patents on Biotechnology, 14(3), 229–234. https://doi.org/10.2174/1872208314666200311104541.
[5] Liu, H., Cao, Y., Guo, J., Xu, X., Long, Q., Song, L., & Xian, M. (2021). Study on the isoprene-producing co-culture system of Synechococcus elongates–escherichia coli through OMICS ANALYSIS. Microbial Cell Factories, 20(1). https://doi.org/10.1186/s12934-020-01498-8.
[6] Team:nevada/Project/Co-cult. (n.d.). Retrieved October 16, 2021, from https://2011.igem.org/Team:Nevada/Project/Co-Cult.
[7] Team:UCL/results. (n.d.). Retrieved October 16, 2021, from https://2017.igem.org/Team:UCL/Results
[8] Towards an Engineered Co-culture Toolbox. Team:Duesseldorf. (n.d.). Retrieved October 12, 2021, from https://2018.igem.org/Team:Duesseldorf.
[9] Ibid.
[10] Ibid.
[11] Team:ITB Indonesia/Results. (n.d.). Retrieved October 16, 2021, from https://2017.igem.org/Team:ITB_Indonesia/Results.
[12] Zhong-Johnson, E. Z., Voigt, C. A., & Sinskey, A. J. (2021). An absorbance method for analysis of enzymatic degradation kinetics of poly(ethylene terephthalate) films. Scientific Reports, 11(1). https://doi.org/10.1038/s41598-020-79031-5.
[13] Ibid.
[14] Mohamed, E. T., Mundhada, H., Landberg, J., Cann, I., Mackie, R. I., Nielsen, A. T., Herrgård, M. J., & Feist, A. M. (2019). Generation of an E. coli platform strain for improved sucrose utilization using adaptive laboratory evolution. Microbial Cell Factories, 18(1). https://doi.org/10.1186/s12934-019-1165-2.
[15] Ibid.
[16] Ibid.
