Hardware

Housing our co-culture system
Our idea of digesting PET plastics comes from the prevalent ocean plastic problem. Focusing on PET, Eco.ly focuses on ‘clean’ biodegradation, i.e. with the least amount of pollution -- and the idea of using biological microorganisms seems ideal. There are already papers showing that bacteria are able to degrade plastics, as early as the research of Ideonella sakaiensis by a team of Japanese researchers back in 2016, but energy input is still needed for the bacteria to perform its function. Therefore, we propose that photosynthetic cyanobacteria can be utilised as a chassis to transform readily available energy into a bacterial energy source, i.e. glucose or sucrose. After confirming that the co-culture mechanism could work, we would like to further investigate its practicality, in particular, how we can actually build a physical system that can contain all the aforementioned processes. A container, or more accurately, a bioreactor, is needed.Initial considerations for Eco.ly bioreactor design
Staying true to our claim of a ‘self-sustaining’ bioreactorOur device makes use of renewable energy to power its functions. Firstly, all the biological and chemical reactions are activated by solar energy, with work input by the microorganisms themselves. As the container is transparent, sunlight can go all the way into the bioreactor and can be used by S. elongatus for its photosynthetic process. S. elongatus would then secrete sucrose as food source for E. coli, which will then secrete PETase and MHETase for degradation of PET. With the design of the filter membranes, the molecules can flow across the concentration gradient into specific compartments of the bioreactor such that the efficiency of the degradation can be maximised, and most likely without the need for extra power source. We are also planning to extract solar-derived energy for the rotary movements of the pulverizer and bioreactor. Efficiency may be compromised with the intensity of sunlight, but heat from some sunlight might be also useful for temperature control since the system works best at a temperature higher than room temperature (~30°C).
Increasing the surface area for reaction
A few modifications are made. First, to increase the surface area for reaction, we would like the PET plastics to be small enough (if the plastic waste is on the scale of a water bottle, it would take too much time to perform degradation. Therefore, we added . Then, for the main bioreactor module, compartments are created by adding specific filter membranes to isolate the bacteria, the cyanobacteria, and enzymes. This helps maintain the concentration of different reactants such that the reaction rate can be kept at a maximum. Furthermore, the arrangement of different components were carefully designed and modified such that it would be simple and convenient to access any components inside (refer to the figure below to see the arrangement and structure of the device).
Feedback controls
After confirming the working mechanism and machine design, we tried to solve more technical problems, mainly proving our system works in a stable manner. Even though results from literature reviews or experiments show that our model could work, we still need a well-planned system control and feedback to make sure our machine works well. Therefore, we looked into the design of a fermentation machine that also involves the growth of microorganisms. We identified several important factors to be taken into account, namely temperature, pressure, pH, O2, and CO2 concentrations, and we also have to monitor the viability of the bacteria from time to time. Therefore, different probes are going to be introduced to the system, together with the feedback response, such that the environment of the bioreactor can be maintained at an optimal condition that favours bacteria growth and enzyme function.
Considerations we took when designing bioreactor’s system controls: Temperature control: It is vital that the temperature of the bioreactor is kept at a certain value such that the bacteria and enzymes inside work stably in the most efficient way. In fact, the optimal working/growth temperature for all components is not the same (30°C for PETase, 37°C for E. coli, and 33°C for S. elongatus PCC 7942), and to prevent death or denaturation of bacteria or enzymes, we would use the lowest temperature among all (i.e. 30°C) even though this means there would be some efficiency trade-off. A temperature sensor is added for providing feedback to regulate temperature inside the bioreactor. In general, a temperature probe is inserted into the container, and after receiving the temperature information, it regulates the system to heat up or cool down. However, when the container is exposed to sunlight for a long period of time, (together with a negligible factor: the metabolic heat generated by the bacteria), the system could go over 30°C. Therefore, a heat jacket with cooling water flow might be needed. Also, since we want a uniform temperature inside the container, stirring might be necessary, and it can be done using the temperature probe / sensor together with the spinning motion of the bioreactor. Stirring is also a method that can alleviate some of the issues mentioned in “Pressure considerations” and “O2 control” below.
Pressure considerations: One of the pressure sources could be due to the spinning of the bioreactor, creating a centrifugal force for solution inside against the container wall. However, since this is a gentle rotary movement, the container should be able to endure that. Respiratory carbon dioxide from bacteria could also create extra pressure. This can be alleviated by spinning and mixing, which helps carbon dioxide dissolve into the solution better. This issue is further addressed in “pH control” and “Monitoring viability of cells”.
O2 control: Apart from sunlight or sucrose as a(n) direct/indirect energy source, bacteria also needs oxygen to grow. Therefore, it is important to measure dissolved oxygen concentration inside the container. Stirring can increase the inflow of oxygen, which introduces air flow into the solution.
pH control: From “Pressure considerations”, we know that CO2 is produced by bacteria during respiration. When CO2 dissolves into water, carbonic acid, a weak acid, is formed. This lowers the pH value of the solution. Also, during the production of protein products (e.g. enzymes), cells would consume ammonia for cellular demand of nitrogen. This will cause proton release and further decrease the pH value. Since bacteria and enzymes have a delicate range of working pH value -- in particular, the pH value of the external environment would affect processes like nutrient uptake (now sucrose uptake by E. coli) and protein secretion (now secretion of PETase and MHETase by E. coli) -- it is important to monitor the pH value in different compartments of the bioreactor, and to adjust it from time to time. Since now there are only factors that would cause pH value to drop, we would need to supply base, for instance ammonia (NH3), as it will transform to basic ammonium hydroxide when it dissolves in water by this chemical reaction: NH3 + H2O <--> NH4+ + OH- <--> NH4OH.
Monitoring viability of cells: Since the bioreactor depends highly on the function of bacteria, we would like to monitor their viability to see if the reaction is maximised. There are various ways to do so. From “Pressure considerations”, we know that measuring carbon dioxide from bacteria’s respiration is one of such methods. This is also known as offgas analysis, a method that can indicate cellular activity. Another common method (which the HKU iGEM team also does for wet lab experiments) is to check the optical density (OD) value, which indicates cell concentration. However, not only does this neglect the effect of ‘dead cells’ in the reactor, this off-line analysis might not be suitable for performing inside a bioreactor. Therefore, “offgas measurement” is more preferred as it can also be used as the pressure sensor.
Semi-permeable membranes
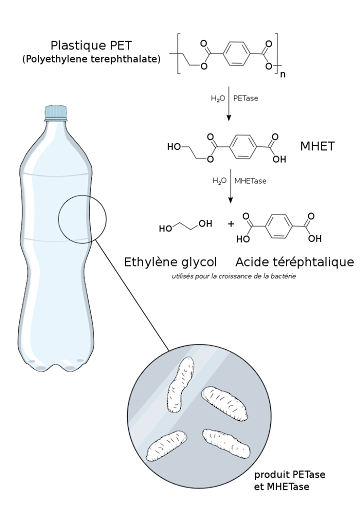
The semipermeable filter is important to separate the innermost region (houses the pulverized PET initially) and the E. coli compartment (together with sucrose originally from S. elongatus) of the bioreactor. The filter would be able to only let the enzymes secreted by E. coli (including PETase and MHETase) to pass through but not E. coli itself, such that the innermost compartment has high enough concentration of enzymes to react with the pulverized plastics.
This filter is made of hydrophobic materials such as polytetrafluoroethylene (PTFE) since the surface of PETase is hydrophobic. A hydrophobic filter also means water cannot pass through, such that there will not be water potential change across the membrane, i.e. PETase can flow through its concentration gradient to the innermost compartment constantly since E. coli keeps on generating the enzymes. A polymer membrane also usually means it is ductile enough to deal with osmotic pressure. Furthermore, sucrose (a hydrophilic molecule) that flows into the middle compartment can hardly go through this PTFE layer, such that E. coli can capture these sucrose molecules easily for consumption and growth. As for the pore size, we have to look into sizes of both PETase and MHETase. PETase exists as a monomer with a crystal structure of the P212121 space group. This is an orthorhombic crystal system where the crystal unit is best enclosed in a cuboid space. Measuring this cuboid space’s 3 dimensions, we find out that the longest dimension of a PETase molecule is around 13 nanometers (PETase’s unit cell parameters: a = 43.48 Å, b = 50.40 Å, and c = 129.49 Å). MHETase is smaller than PETase (as it can also be induced since MHETase digests MHET, a much smaller molecule than PET, which is digested by PETase; the pathway can be referred to the pathway figure below). Hence any filter with pore size between the range of tens of nanometers to micrometer scale (typical size of E. coli) would suffice.
Designing the bioreactor
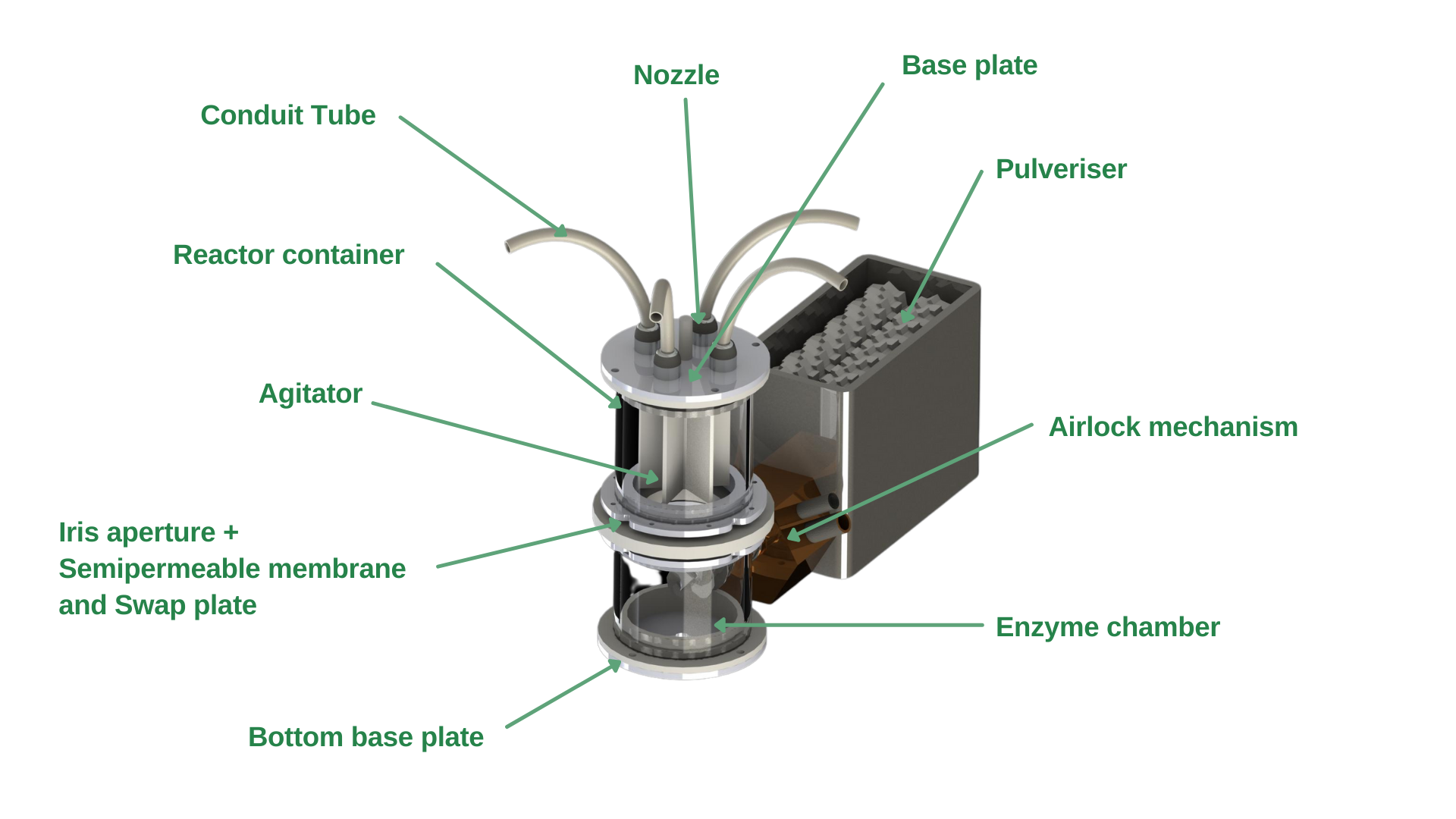
Referring to Figure 1, the container’s design consists of a pulveriser, an airlock mechanism, and a main body composed of a co-culture chamber, enzyme chamber, inlet tubes, a shutter and a semi-permeable membrane.
The process is as follows:
1. The co-culture system of E.coli and cyanobacteria is contained within the co-culture chamber (pictured in Figure 2). This chamber is transparent, allowing light necessary for cyanobacteria photosynthesis to pass through. This chamber also contains an agitator (stirrer), which ensures the organisms, temperature and substrates (e.g. oxygen and sucrose) are evenly distributed within the co-culture to maximise reaction rates. The inlet tubes allow for input of probes to monitor the conditions within the co-culture system, as well as provide substrates if necessary to maintain the co-culture.
2. The enzymes produced by E.coli passes through the semipermeable membrane into the enzyme chamber.
3. For PET digestion, the pulverizer (pictured in Figure 3) will first grind the PET into smaller pieces.
4. The PET pieces are transferred to the enzyme chamber through the airlock component via the screw impeller (pictured in Figure 4). The airlock component is necessary to prevent the backflow of liquid from the enzyme chamber due to the difference in pressure between the two phases.
5. In the enzyme chamber, the PET pieces are mixed with the enzymes (PETase and MHETase) produced by E.coli. The enzyme chamber is detachable from the co-culture chamber due to the presence of the iris aperture (pictured in Figure 5). The aperture is shut before the disconnection of the enzyme chamber, allowing us to separate the enzyme chamber for monomer collection without any spillage of liquid from the co-culture chamber.




Mechanical Design Innovations
The aperture-closing is based on the shutter design of a camera, allowing for the easy opening and closing of the aperture while maintaining a compact form factor. This allows us to modularize the system and to easily extend the capacity of the bioreactor chambers.Second is the 'airlock' mechanism, which allows a continuous form of crushed PET transport into the enzyme, allowing for a seamless process from PET bottle to monomers. This is coupled with the impeller system which functions as an Archimedes screw and an agitator for the enzymatic reactions. The aperture-closing is based on the shutter design of a camera, allowing for the easy opening and closing of the aperture while maintaining a compact form factor.
Material Choice
Stainless steel is chosen for the structural components for the reactor as it has good strength properties. It is also corrosion-resistant. Food safe PETG is chosen for the auxiliary parts of the reactor. Please refer to the bill of materials for each part's material specification.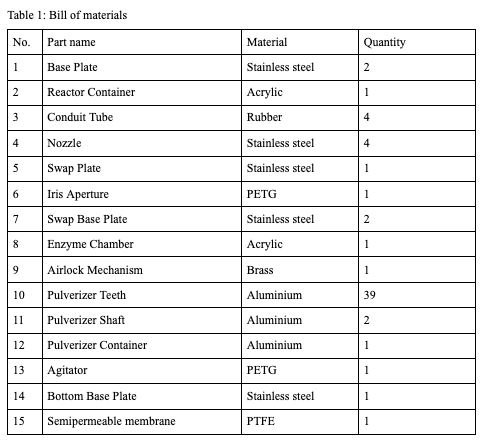
References
[1] Kawai, F. (2021). The Current State of Research on PET Hydrolyzing Enzymes Available for Biorecycling. Catalysts, [online] 11(2), p.206. Available at: https://www.mdpi.com/2073-4344/11/2/206/htm[2] Rashed Noor, Zahidul Islam, Saurab Kishore Munshi and Farjana Rahman (2013). Influence of Temperature on Escherichia coli Growth in Different Culture Media. [online] ResearchGate. Available at: https://www.researchgate.net/publication/233761514_Influence_of_Temperature_on_Escherichia_coli_Growth_in_Different_Culture_Media
[3]Kuan, D., Duff, S., Posarac, D. and Bi, X. (2015). Growth optimization ofSynechococcus elongatusPCC7942 in lab flasks and a 2-D photobioreactor. The Canadian Journal of Chemical Engineering, [online] 93(4), pp.640–647. Available at: https://onlinelibrary.wiley.com/doi/full/10.1002/cjce.22154
[4] Cytiva. (2021). [online] Cytiva. Available at: https://www.cytivalifesciences.com/en/us/solutions/lab-filtration/knowledge-center/membrane-filtration-choosing-the-correct-type-of-filter
[5]Joo, S., Cho, I.J., Seo, H., Son, H.F., Sagong, H.-Y., Shin, T.J., Choi, S.Y., Lee, S.Y. and Kim, K.-J. (2018). Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nature Communications, [online] 9(1). Available at: https://www.nature.com/articles/s41467-018-02881-1
[6] Cytiva. (2021). [online] Cytiva. Available at: https://www.cytivalifesciences.com/en/us/solutions/lab-filtration/knowledge-center/membrane-filtration-choosing-the-correct-type-of-filter
[7] Palm, G.J., Reisky, L., Böttcher, D., Müller, H., Michels, E.A.P., Walczak, M.C., Berndt, L., Weiss, M.S., Bornscheuer, U.T. and Weber, G. (2019). Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nature Communications, [online] 10(1). Available at: https://www.nature.com/articles/s41467-019-09326-3
[7] Wikipedia Contributors (2021). PETase. [online] Wikipedia. Available at: https://en.wikipedia.org/wiki/PETase#/media/File:Plastic_Breakdown_by_PETase.png
