Overview
Our project aims to genetically engineer a naturally transformable lab strain of Escherichia coli to produce a more affordable and efficient bacterial host for synthetic biology. Although E. coli is the preferred host for synthetic biology, it is not naturally competent, and requires treatment to become chemically or electrocompetent for effficient transformation. Genes from the related non-pathogenic and naturally competent Acinetobacter baylyi will be integrated into E. coli through recombineering. A. baylyi's natural transformation and competence genes will be inserted as eight separate gBlocks and integrated sequentially with an alternative selection markers.
Our Inspiration
The power to harness synthetic biology to better our planet lies in the hands of those with access to research infrastructure, expertise and a skilled workforce. Our team was inspired to tackle this inequity to ensure that synthetic biology remains a tool to better the world, rather than a tool to increase inequality. Our design for a naturally transformable lab strain of Escherichia coli - Free Coli - is here to save the day by making affordable, efficient and safe synthetic biology education and research accessible for all.
Our Solution
We have designed a series of parts and experiments to engineer this naturally transformable host organism that can take up and integrate foreign DNA without the need for chemical treatment or electroporation to become competent. Our team consulted with bacterial competence experts and reviewed the literature on natural transformation in E. coli and related bacteria to develop our approach and identify twenty-three putative natural transformation genes from the related, non-pathogenic gram negative bacterium Acinetobacter baylyi.
The literature revealed that while E. coli possesses homologs for almost all of A. baylyi's natural transformation genes, the genes were not functioning as observed in A. baylyi and E. coli's endogenous natural transformation promoter could not be successfully induced to produce a naturally transformable phenotype. This informed our decision to 'import' A. baylyi's natural transformation genes into the JM109 E. coli lab strain.
Due to the high number of genes predicted to be necessary to produce a naturally transformable strain, our team devised a novel recombineering strategy for insertion of multiple gene clusters - Babushka Blocks! We were limited by the maximum size of DNA able to be commercially synthesised into a gBlock - 5kb. Our modelling team employed k-means clustering and existing data on transcriptome concentration and promoter strength to model the optimal clustering of genes into eight < 5kb DNA fragments. Bioinformatics analysis was conducted to assemble the genes into fragments with salicylate promoters and selectable markers. Synthetic promoters were used as part of our Free Coli design as a safety feature and to improve efficiency - enabling researchers to place the natural transformation ability of the strain under the control of a simple inducible promoter. Our 'Babushka Blocks' novel recombineering strategy also integrated key learnings from our consultation with antibiotic resistance experts to minimise the number of antibiotic resistance genes carried by our end-product. The strategy sees each subsequent gene cluster insert within the protein coding sequence of the previous cluster's antibiotic resistance gene - effectively knocking out the previous resistance gene so that Free Coli only ever contains one antibiotic resistance gene, both during the design phase and as a final product for researchers.
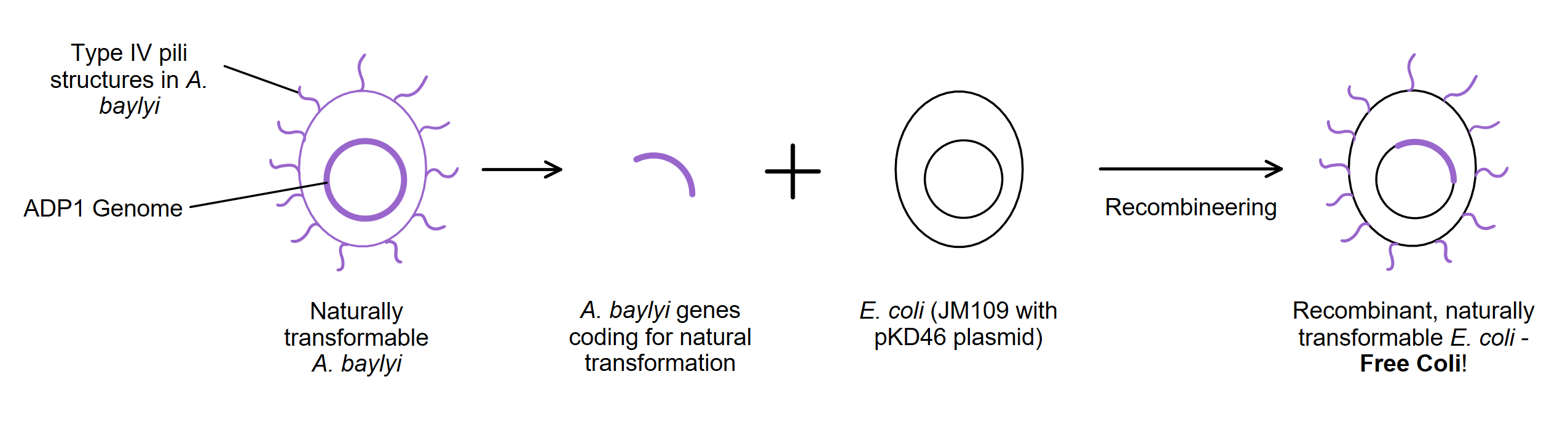
Figure 1. Simple diagram describing the construction of Free Coli. 23 genes responsible for natural transformation in A. baylyi will be synthesised and recombineered into a strain of E. coli. This will yield a strain of E. coli that does not need expensive chemical or electrical treatments to induce competency.
Why is Our Project a Useful Application of Synthetic Biology?
Throughout the course of our project, the wider community was engaged and consulted to affirm the project's purpose and align its future implementation with the UN's Sustainable Development Goal to advance equality around the world. Our design for a naturally transformable strain of E. coli is an example of a foundational technology that could be applied to break through the barriers preventing the establishment of thriving research ecosystems in developing countries. Free Coli can therefore advance multiple SDG goals by contributing to the establishment of a research and development capability and underpinning the economic, health, agricultural and environmental benefits of translating and commercialising synthetic biology research.
How COVID-19 Affected Our Project
Due to the impact of COVID-19 outbreaks and lockdowns in Sydney during 2021, our team was unable to gain access to our laboratory to carry out our experiments and validate our design. While we initially planned to validate our hypothesis and design in a series of wet lab bacterial transformation experiments using our novel recombineering strategy for multiple gene cluster insertions, the Sydney lockdown required the team to pivot mid-project and focus on strengthening our design through modelling. We hope future iGEM teams take our research, comprehensive design and novel experimental strategy to assay engineering success into the lab for the first time to validate our design and turn our idea into reality.
In concert with the accessible synthetic biology technology we researched and designed, we identified a need to improve and expand the pipeline of highly skilled synthetic biology researchers to fulfil future demand, and thus pivoted our project by increasing our focus on education and outreach. We believe that shaping the scientists of tomorrow starts by sowing the seeds of scientific curiosity in children. A key focus for our project was to engage with primary and high school students to teach and spark their curiosity in synthetic biology. We prepared educational materials that demonstrated the concepts underpinning our project and aligned them to the curriculum for primary school students and high school students studying biology from Years 9 to 12. Our resident musician Rhys also recorded several educational songs parodying rock classics to teach foundational synthetic biology concepts in a fun and engaging way.





