OVERVIEW
The ChiSPY biosensor will detect the presence of the chitin oligosaccharide N-acetylglucosamine by expressing the chitin-sensing one-component system from Vibrio cholerae [2,3] in E. coli coupled with a GFP reporter. The presence of the chitin oligosaccharide is indicative of chytrid infection caused by either Batrachochytrium dendrobatidis (Bd) or Batrachochytrium salamandrivorans (Bsal) [5] due to the life cycle of the chytrid infection. Chytridiomycosis has caused the decline of at least 500 species of frogs and salamanders [7]. Due to the massive decline, Lubbock_TTU decided to help!
INSPIRATION
In spring 2020, the Lubbock_TTU team spoke to our PI, Dr. Michael San Francisco, regarding various projects that warranted further investigation. He introduced us to the chytrid fungal pandemic (Bd) and it caught the attention of the team because of how widespread, yet unknown it is. Over these past two years, we continued to investigate the Bd pandemic and major issues related to it. We saw the potential for a project regarding this infection because we could see how synbio could easily fit into two current mitigation methods: immunization and treatment of host and habitat [4]. Due to what we thought was a clear idea of how synbio could fit into those two mitigation techniques, we planned to do a bioaugmentation project.
One of our first goals in the bioaugmentation project was just to have the amphibian live long enough to reproduce. If the amphibian was able to reproduce then species on the decline would be able to increase their population. We looked into areas where an engineered microbe could benefit the host, damage the fungus, and/or offset the effects of the fungus long enough for the amphibian to reproduce. We looked into the native microbiome of frogs, areas in the world where Bd is more virulent, climates where the infection rate was higher than most, and what species of frogs were more susceptible to the infection[4]. As we continued to gather other options and learn more about the Bd pandemic, we came across 2 experts, Dr. David Rodriguez and Dr. Ankur Dalia, and consulted with them..
When we first met with Dr. Rodriguez we originally wanted to ask if he knew of any reason why certain species of frogs are more resistant to Bd . However, during that same meeting, he emphasized the lack of available and cheap equipment to detect the presence of Bd on amphibians. Current detecting equipment such as MinION ($1,000-5,000) and MicPCR ($16,000-20,000). These prices do not include the environmentally sensitive reagents, consumables, and prep kits required to maintain the use of these such equipment. This new information made the team realize there was something greater we could do to help mitigate this pandemic-create an affordable detection method.
Since our focus was now on detecting Bd , we began to look into what was already being done to detect it. This included looking into past iGEM biosensors (2017 UFlorida and 2020 UMaryland teams), biomarkers we could use to identify the presence of the infection, and other field ecologists who identify regions of interest. Biomarkers that we looked into include eDNA, specific proteins, and secondary metabolites. One issue we ran into while searching for potential biomarkers was the copy number variation. In Bd’s genome, the ITS1 region is copied in different amounts [6] . The ITS1 region is present in all genomic biomarkers, and it makes it very difficult to determine if one strain of Bd is more virulent than the other. To negate this problem, we decided not to use a genetic biomarker. This eliminated eDNA as a viable option for us.
The biomarker we decided to use is chitin. We came to this conclusion because the sporangia shells are made of chitin and we concluded that if there were sporangia shells then there would be infection [5]. Once we figured out which biomarker we wanted to use, we began looking into sensing pathways to detect this oligosaccharide.
One method of detection we found was a two-component histidine kinase [3]. The reason we decided not to use this is because we discovered the V. cholerae one-component system, which is a noncanonical hybrid sensor kinase[3]. Dr. Ankur Dalia contributed to the development of our project by sharing possible genes to include in our Bd biosensor construct such as ChiS, Pchb, and CBP. We continued to brainstorm ideas that led us to center around a chitin biosensor project for this year's competition. Finding this detection method was a major breakthrough for the team because it gave us an even clearer idea of what we could do with this project.
Because this pandemic is spread across globally, we wanted to ensure that places across the world had something they could use to detect it. With this thought in mind, we entered what would be our final project idea.
PROJECT DESCRIPTION
The Bd Lifecycle
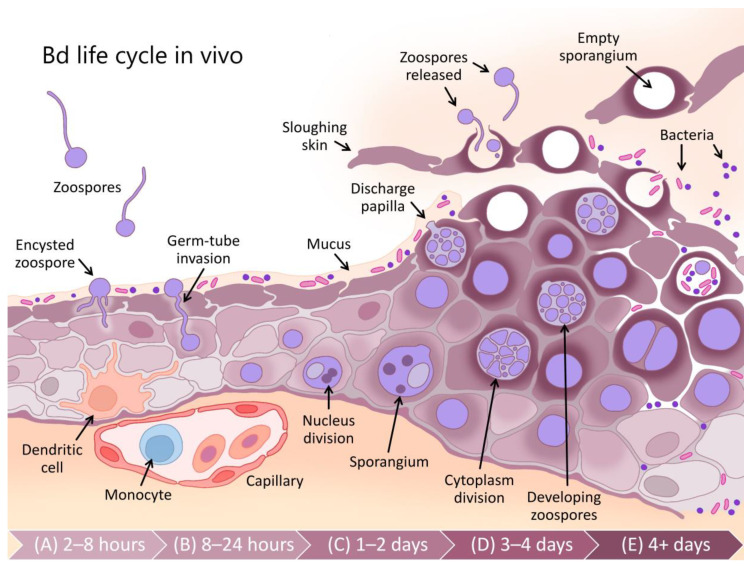
Bd is the causative fungus of chytrid disease. Figure 1 shows how the fungus spreads through zoospores that encyst into epidermal tissue via germ-tube invasion. After the zoospores have invaded the dendritic cells, they undergo nuclear division and develop into zoosporangia. These zoosporangia have cell walls that contain chitin–the biomarker of our system–and undergo cytoplasmic division that engender many zoospores [5]. As the amphibian skin gets sloughed off, the developing zoosporangium gets closer to the surface of the epidermis and releases the internal zoospores through discharge papillae[1]. These zoospores are now free to spread to other amphibians and continue the Bd infection cycle.
The Vibrio cholerae Chitin Utilization Program
The Vibrio cholerae bacteria is the causative agent of the diarrheal illness cholera. This pathogen creates biofilms on the chitinous shells of microscopic crustaceans to persist in the environment, transmit to hosts, and evolve. The chb operon (genes responsible for degradation, uptake, and utilization of chitin in V. cholerae ) is directly regulated by the one-component hybrid sensor kinase, ChiS, differing from prototypical two-component signal transduction systems [2]. When chitin is not present in the periplasm, ChiS activity is repressed by chitin binding protein (CBP). When chitin is present as oligosaccharides after being digested by chitinases in the extracellular space, it binds to CBP and relieves repression, allowing ChiS to bind to the promoter and activate expression of chb [3] . The nucleoid occlusion protein SlmA also activates the chb operon by directly binding to its promoter. [3]

Chitin-Sensing Pathway (ChiSPY) Biosensor
As the formation of zoosporangia indicates reproduction of zoospores and thus infection, chitin– a polysaccharide found in the sporangium cell wall– will serve as the biomarker for detection of chytrid infection. This biomarker deviates from traditional detection methods (qPCR) that detect zoospore presence. qPCR cannot determine infection intensity as it relies on the ITS1 region of the Bd genome to indicate fungal presence [6].This region has variable copy numbers across different strains, making qPCR an unreliable diagnostic tool for determining infection intensity. To detect the prevalence of zoosporangia and thus infection intensity, we aim to create a live cell biosensor by expressing the V. cholerae one-component chitin-sensing system coupled with a GFP reporter in E. coli.
Chitin oligosaccharides will be taken up by the cell through chitoporins in E. coli’s outer membrane. Once in the periplasm, the oligosaccharides bind to CBP and inhibit phosphorylation of integral protein ChiS, allowing activation of GFP expression. The ChiSPY biosensor serves as a cheap initial detection method to identify potential “hotspot” regions of amphibian infection, prompting further investigation by ecologists. This tool would be especially useful in areas where expensive tools like qPCR or MinION are not easily accessible. Additionally, using a live cell biosensor allows better quantification of the varying levels of fluorescence that reflect different infection intensities.

Future Outlook
The ChiSPY biosensor will produce GFP when induced by chitin oligosaccharide prevalence. Chitin-sensing biosensors have other detection applications. For example, the ChiSPY biosensor can be used to detect fungal infections in humans, or as a marker for antifungal susceptibility tests. In terms of the chytrid pandemic, there is potential to apply the ChiSPY biosensing circuit directly to the amphibian microbiome for in situ monitoring. Additionally, the biosensor could be coupled with a circuit that produces antifungal metabolites or probiotics that directly treat the infection.
Sources
[1] Grogan LF, Humphries JE, Robert J, Lanctôt CM, Nock CJ, Newell DA, McCallum HI. Immunological Aspects of Chytridiomycosis. J Fungi (Basel). 2020 Oct 19;6(4):234. doi: 10.3390/jof6040234. PMID: 33086692; PMCID: PMC7712659.
[2] Klancher, Catherine A., et al. “ChiS Is a Noncanonical DNA-Binding Hybrid SENSOR Kinase That DIRECTLY Regulates the CHITIN Utilization Program in Vibrio Cholerae.” PNAS, National Academy of Sciences, 18 Aug. 2020, www.pnas.org/content/117/33/20180.
[3] Klancher, Catherine A., et al. “The Nucleoid Occlusion Protein SlmA Is a Direct Transcriptional Activator Of Chitobiose Utilization in Vibrio Cholerae.” PLOS Genetics, Public Library of Science, 6 July 2017, journals.plos.org/plosgenetics/article?id=10.1371%2Fjournal.pgen.1006877.
[4] Woodhams, Douglas C, et al. “Mitigating Amphibian Disease: Strategies to Maintain Wild Populations and Control Chytridiomycosis.” Frontiers in Zoology, BioMed Central, 18 Apr. 2011, frontiersinzoology.biomedcentral.com/articles/10.1186/1742-9994-8-8.
[5] JE;, Berger L;Hyatt AD;Speare R;Longcore. “Life Cycle Stages of the Amphibian Chytrid Batrachochytrium Dendrobatidis.” Diseases of Aquatic Organisms, U.S. National Library of Medicine, Dec. 2005, pubmed.ncbi.nlm.nih.gov/16465834/.
[6] Longo, Ana V., et al. “ITS1 Copy Number Varies among Batrachochytrium Dendrobatidis Strains: Implications for QPCR Estimates of Infection Intensity from Field-Collected Amphibian Skin Swabs.” PLOS ONE, Public Library of Science, https://journals.plos.org/plosone/article?id=10.1371%2Fjournal.pone.0059499.
[7] Wildlife Futures Tea. “Chytridiomycosis (BD).” PennVet.com, 21 Mar. 2020, https://www.vet.upenn.edu/research/centers-laboratories/research-initiatives/wildlife-futures-program/resources/fact-sheets/fact-sheet-detail/chytridiomycosis-(bd)#:~:text=Over%20700%20species%20of%20amphibian,et%20al.%2C%202019).
