
BLADEN
Design
Phased approach
Here we describe the general overview of our project in terms of design on the wet lab front. We effectively split our project into three main phases to reach our final goal of adapting EvolvR in planta:
Phase 1: Testing EvolvR in bacteria
In order for us to introduce EvolvR into a new biological system, we sought to confirm the efficiency and functionality of EvolvR in bacteria as previously described [1]:

Phase 2: Testing EvolvR in a cell-free system made from plant cell lysate
Before testing EvolvR directly in plant cells, we wanted to validate its functionality in a eukaryotic cell environment by testing it in a cell-free system made from Tobacco BY-2 cell lysate:

Phase 3: Incorporation of a functional EvolvR in BY-2 cells
Our ultimate goal being to validate that EvolvR can function as expected in BY-2 plant cells, we established a rapid method for a continuous directed evolution experiment with BY-2 cells:

Additional experimental design
Molecular ruler experiment
To measure the editing window of EvolvR, we designed ten evenly distributed protospacers throughout the GFP gene:

Riboswitch experiment
Because we do not have an inducible system in plants for EvolvR, we believed that evolving a riboswitch could be an avenue for us to implement such a system in a plant vector.

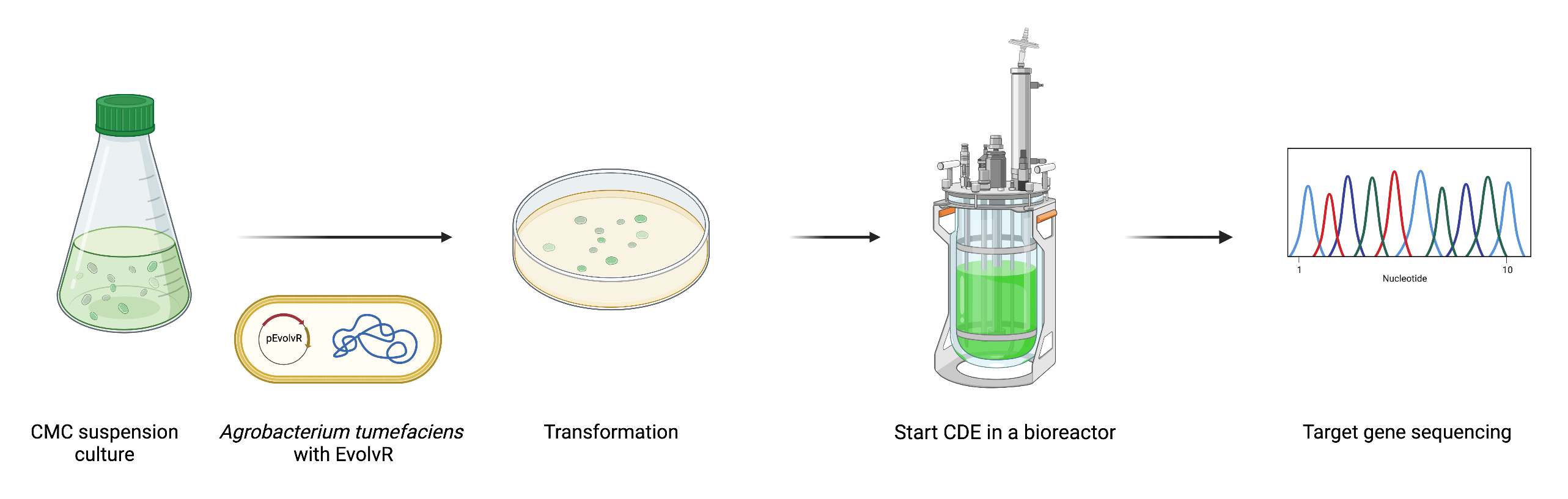
Establishment of a CMC suspension culture
Beyond the BY-2 suspension culture, we believe that establishing a suspension culture from CMCs, plant stem cells, will be beneficial for the implementation of EvolvR in plant biotechnology research.
References
[1] Halperin, S. O., Tou, C. J., Wong, E. B., Modavi, C., Schaffer, D. V., & Dueber, J. E. (2018). CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature, 560(7717), 248–252. https://doi.org/10.1038/s41586-018-0384-8

