
BLADEN
Project description
Introduction
Due to climate change, local conditions around the world are changing rapidly. The weather is becoming more unpredictable with large and sudden deviations from stable local conditions. The effects of climate change have a particularly large impact on agriculture and farmers must adapt their crops to these new conditions. Plants are versatile organisms that have an extensive secondary metabolism that allows them to respond to biotic and abiotic stresses, but they cannot adapt and evolve at the same pace as the climate is changing, at least not through classical breeding techniques or on their own.
Fortunately, the field of biotechnology is rapidly advancing. New genomic techniques are constantly being developed to easily genetically modify plants. These advances open many new roads for the development of more sustainable agriculture using genetically modified organisms (GMOs). Many of the stresses related to climate change, such as drought and heat stress, can be addressed by creating new GMOs. Nowadays, most GMOs are engineered by using rational design that needs established biological knowledge. However, the field of biology is immensely vast, and it is impossible to study every biological process in detail. This becomes more difficult when the biological system that needs to be altered by genetic modification tools becomes more and more complex.
Our team is trying to circumvent this by developing a semi-rational design method for GMO crop engineering — we are going as far as only needing sequence data and very approximate structures of the proteins that are being targeted. The goal is to evolve and improve plants at a faster pace through this semi-rational design using Continuous Directed Evolution (CDE).
Directed Evolution
Directed evolution (DE) is a well-established laboratory method that optimizes proteins for a target environment using Darwinian logic.
Principles of Directed Evolution
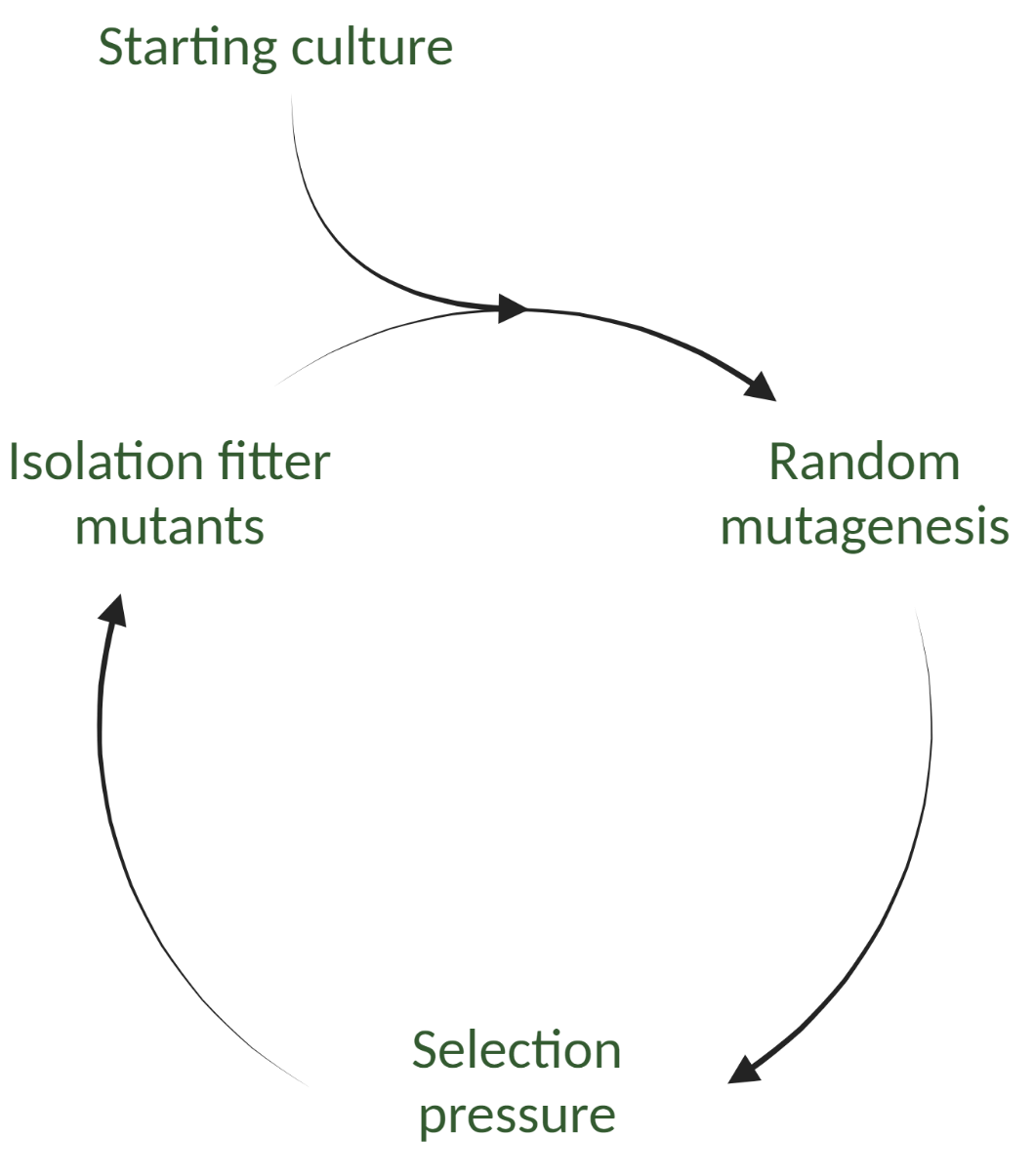
In the first step, different random mutations are made in the genomic DNA of organisms or cells to create a library of mutants. Mutations are made via random mutagenesis, which randomly changes the nucleotide sequence of the target gene.
The performance of each mutant is then screened using a desired selection method, and mutants with improved characteristics can be isolated. Another round of DE may be started using these isolated mutants. This process continues until the desired performance increase is reached in whatever trait is being targeted, which usually takes 4-6 rounds of DE. Figure 1 shows an overview of the directed evolution process.
Challenges for DE in plants
Mutant production and growth
Performing DE on unicellular organisms is relatively easy as thousands of mutants can be generated in high-throughput due to short generation times and minimal biomass production per individual. However, the generation of mutant plants for the first step of a DE experiment can often take more than 6 weeks. This is due to the fact that a multicellular organism, such as a plant, needs time to grow and develop.

Transformational efficiency
Most mechanisms are limited by transformation efficiencies, which measure the efficiency by which a living cell can take up extracellular DNA and express the encoded genes. Transformation remains challenging for many organisms, including plants. As this is a limiting factor in traditional DE, this approach remains valuable, but not a perfect tool for crop improvement specifically. In our project, we propose a novel and rapid approach based on enzymatic mutagenesis in plant cell culture.
Project proposal
Mutational strategy using EvolvR
A new in vivo mutational technique has been developed called EvolvR that allows for continuous directed evolution [2]. EvolvR relies on a mechanism that is based on the bacterial defense system called Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) in which CRISPR sequences are recognized and cut by CRISPR-associated (Cas) enzymes, as shown in Figure 3. EvolvR uses a nicking variant of a Cas enzyme that creates single-stranded nicks in the DNA, and that is fused to an error-prone DNA polymerase. As the Cas enzyme binds to a target region via a guide RNA, it nicks the DNA, which allows for the error-prone DNA polymerase to bind and synthesize a new strand with errors while displacing and degrading the original strand. As long as EvolvR is expressed within the cells, this process of nicking and error-prone strand synthesis can continue, thereby allowing for CDE at a target locus.

Our goal is to introduce the EvolvR mechanism in plant cells to perform CDE. The advantage of continuous over ‘traditional’ directed evolution in plants is that it doesn’t require multiple rounds of selection and isolation of mutants. Instead, DE can occur continuously in a growing plant cell under evolutionary pressure in a selective environment. We are using Agrobacterium-mediated transformation of a Tobacco Bright Yellow 2 (BY-2) cell line grown in liquid culture to introduce DNA containing EvolvR and a gRNA for the target gene of interest. The DNA is incorporated into the genome of the BY-2 cells which are then subjected to selective pressure.
Advantages
- EvolvR allows to target a specific, user-defined region in a gene to be mutated.
- EvolvR allows for all bases to be mutated (with a little bias towards A and T).
- EvolvR allows us to perform CDE instead of time-consuming traditional directed evolution in cycles.
Disadvantages
- The mechanism still requires transformation of BY-2 cells at the start of a CDE experiment, thus relying on transformation efficiency at the first stage of the experiment.
- EvolvR has been tested in yeast, but not yet in plants [2].
- BY-2 liquid cell culture is not uniform, which may make isolation of single mutants more difficult, and the growth rate is still relatively low, slowing down the acquisition of desired results.
Shortening generation time using cambial meristematic cells (CMCs)
CMCs are cells derived from the procambium of a plant, the tissue from which primary vascular tissues develop. They are undifferentiated cells that grow indefinitely and function as plant stem cells. They are responsible for the production of xylem, which transports water and minerals from the roots to the shoots, and phloem, which mainly transports sugars and other nutrients from leaves for storage and towards growing parts of the plant. CMCs are characterized as fast and uniform growing single cells which maintain their morphological and physiological stability.
Advantage of using CMCs
- Easier to transform due to CMCs’ single-celled nature.
- Shorter generation time allowing for faster results from CDE experiments.
Disadvantages
- By using single cells, we could not perform CDE on actual physiological structures of a plant. For example, we cannot alter stomatal function in plant leaves to limit water loss. However, we can optimize almost any metabolically relevant protein in these plant cells.
Proof of concept: Herbicide resistance
We are using herbicide resistance as an easy readout of our system. We can easily test to see if plants are resistant to a particular herbicide by determining whether they have survived in the selective medium or not. Find out more about our approach on the Proof-of-Concept page.
Phased approach
Plant research can be slow and difficult due to the complexity of plant systems. Therefore, we are preparing for potential obstacles by introducing different phases in our project. These phases effectively divide our project into multiple sub-projects that can be organized, performed and analyzed in parallel:
Phase 1: Testing EvolvR in bacteria
- Goal: We will test EvolvR variants in bacteria to confirm that they function as previously described by Halperin and colleagues (2018) [1].
- Use: It will confirm whether EvolvR functions as expected in order for us to apply it to a new biological system.
Phase 2: Testing EvolvR in a cell-free system made from plant cell lysate
- Goal: We will test EvolvR variants in this system in parallel to BY-2 cells as results can be obtained much more rapidly.
- Use: It will give us an indication of the functionality of EvolvR variants in a plant/eukaryotic context.
Phase 3: Incorporate a functional EvolvR in BY-2 cells
- Goal: We have selected BY-2 cells as our model system as they grow at a fast rate in liquid culture and are relatively easy to manipulate. BY-2 cells are then transformed with an EvolvR/gRNA pair. Several EvolvR variants will be tested for their performance in BY-2 cells.
- Use: We will perform proof-of-concept CDE experiments using BY-2 cells. These experiments will serve to quantify, sequence and analyze the efficacy of EvolvR activity in BY-2 cells.
Phase 4: Cambial meristematic cell (CMC) suspension culture
- Goal: We create a stable suspension culture of CMCs of a plant of our choice to incorporate EvolvR into.
- Use: CMC cultures can be used to mass produce certain plant-derived products. Therefore, we should choose a plant which produces a desirable product naturally.
Expected outcome
By testing EvolvR in plant cells, our goal is to make it easier to perform CDE of plant-specific proteins. We want to show that:
- EvolvR remains functional in both an in vitro cell-free system and in vivo in BY-2 cells.
- EvolvR can produce mutants that increase plant fitness in a particular selective context.
- CMC cells can be grown as a suspension culture that can accommodate CDE experiments with EvolvR.
Impact of Covid-19 on our Project
The effect of Covid-19 on our project has been significant. At the start of the project, we could not meet in person. We limited ourselves to online meetings on Zoom. Once the rules relaxed, we organized walks with different team members to brainstorm in subgroups. Although online meetings are not the most conventional way to brainstorm, much less meet each other, we tried to make the best of this situation. At the time, the safety of all the members was of utmost importance.
However, this has had its downsides. Due to the online nature of things, we did find it difficult to brainstorm for long hours. These missed discussions ended up being missed opportunities for us. We also could not elaborate on specific details of the project, which then had to be done at a later stage, creating a stressful scenario.
During most of the last year in Belgium, it was not possible for up to 15 people to meet. So the whole team got together only at the last stage of the project. This significantly affected our team morale. Due to strict measures at KU Leuven, Covid-19 also affected our lab work. A limited number of people could work at the lab at a given time.
References
[1] Halperin, S. O., Tou, C. J., Wong, E. B., Modavi, C., Schaffer, D. V., & Dueber, J. E. (2018). CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature, 560(7717), 248–252. https://doi.org/10.1038/s41586-018-0384-8
[2] Tou, C. J., Schaffer, D. V., & Dueber, J. E. (2020). Targeted diversification in the S. cerevisiae genome with CRISPR-guided DNA polymerase I. ACS Synthetic Biology, 9(7), 1911–1916. https://doi.org/10.1021/acssynbio.0c00149

