

Proof Of Concept
Overview
In this section, our team has proved the validity of the concept of our project and diagnostic kit GotCha. Though we were greatly restricted in terms of lab arrangements, we still did our best in doing as many verifications we could with the limited time. For prior information on the design of our experiments, please refer to Experiment Design.
Circularisation of Linear DNA
Circular probe was synthesized by ligation of the linear DNA. 30 μL reaction solution containing 1 μM linear DNA, 1 μM ligation primer, T4 DNA ligase buffer and 10U/ μL T4 DNA ligase was incubated at room temperature for 4.5 h. Then, linear DNA not circularised and ligation primers were digested by adding Exonuclease I and Exonuclease III. Finally, the two exonuclease were denaturated at 80 °C for 20 min.
We took the circularized DNA to run gel-electrophoresis, and evaluated the result of DNA ligation.
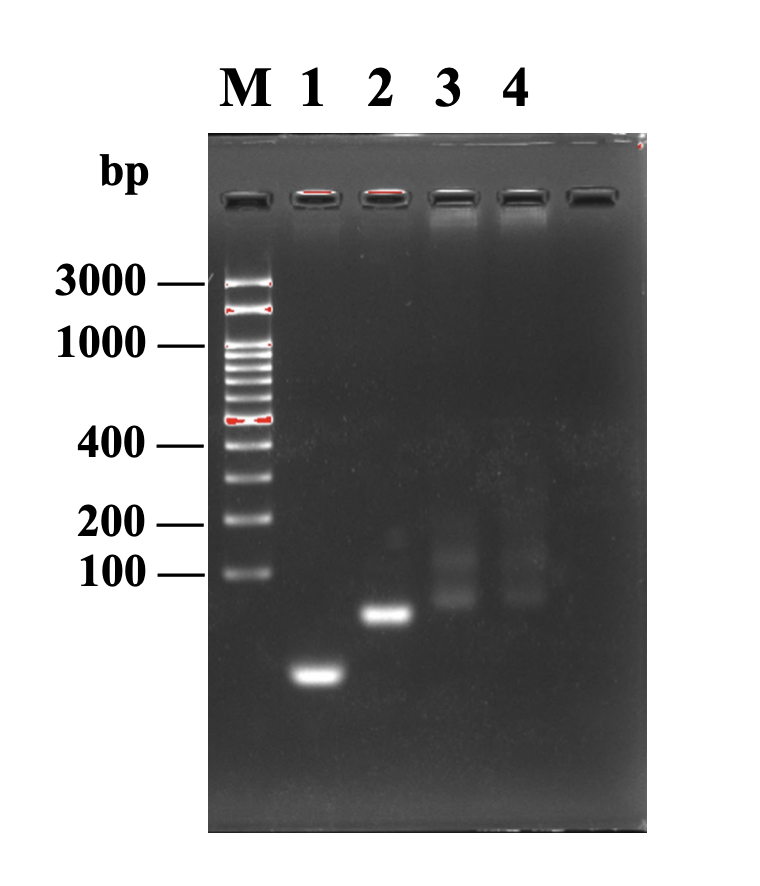
Fig.1
Circularized DNA were analyzed by agarose gel electrophoresis. M: marker, 1:T4 ligase primer, 2: linear DNA, 3: CPB-664a-3p, 4: CPB-33b-3p
The gel showed a “smear” in the lane of circular DNA probe after ligation, which is a common observation in circular DNA gel-electrophoresis due to its conformation.
However, we are aware that the "smear" cannot confirm successful circularisation of linear DNA, as there is a possibility of contamination as well.
RCA action and Linear DNA Circularisation
As we have only used our software model (for more information, please refer to Model) to design our circular probe sequence, in this segment of our experiment, we attempted to verify binding of target miRNA to our circular probe, as well as circularisation of our Circular Probe.

Fig.2
Verification of Circular probe 664a-3p. ***p < 0.001: Ctrl w/o miRNA compared to miRNA 664a-3p. ***p < 0.001: Ctrl w/o phi29 compared to miRNA 664a-3p. CBP: Circular Probe

Fig.3
Verification of Circular probe 33b-3p. ***p < 0.001: Ctrl w/o miRNA compared to miRNA 33b-3p. ***p < 0.001: Ctrl w/o phi29 compared to miRNA 33b-3p. CBP: Circular Probe
As seen in the above graph, it can be interpreted that our test with the RCA reaction mixture was successful in carrying out the desired mechanism.
Comparing the test with RCA reaction mixture to the control without phi29 polymerase in the mixture, it can be seen through the absorbance value that the presence of phi29 polymerase has greatly reduced DNA product output significantly.
From the above, we can establish the following:
- As the magnitudes of difference between the test with the RCA mixture and both controls are very significant, it can be concluded that RCA reaction was indeed carried out.
- For the RCA reaction to be carried out, it can be further concluded that the linear DNA sequence of our Circular DNA Probe was successfully circularised into a Circular DNA Probe and that the target miR has bound to our Circular DNA Probe to act as a primer to start RCA.
Phi29 polymerase action and Optimal Reaction Time
We have also conducted RCA experiments at different durations.

Fig.4

Fig.5
From the graphs above, it can be seen that there is an upwards trend from 0.5h to 2h. From the above information, we can deduce that there was indeed phi29 polymerase elongation reaction over time, eliminating the possibility of absorbance value from other factors other than phi29 action in RCA.
However, it is also to be noted that at the 3h mark, there is an observed decrease in absorbance compared to before. This suggests degradation of DNA product from RCA.
Therefore, we have decided to treat 2h as the ideal duration for our kit for highest efficiency.
RCA action against miR concentration
Finally, we attempted to validate the concept of our mechanism by testing different concentrations of target miRs to derive fluorescence intensity.

Fig.6

Fig.7
As seen above, it can be seen that there is an increase in fluorescence absorbance with increase in miR concentration in both cases.
From here, we can that given the above two verification tests, it can be said that an increase in target miR concentrations led to an increase in amount of product DNA.
We are well aware that our experiment result here is not sufficient enough of a set of data to completely verify the validity of a trend in our kit. However, due to the restrictions imposed by the covid-19 pandemic as well as time constraints, we were unable to do further tests on more concentrations of miR.
Function of GotCha
To implement the idea of GotCha, we conducted several experiments. Through hybridization and immobilization, we successfully assemble GotCha. In next step, we designed a principle-of-proof experiment to verify the feasibility of the GotCha for miRNAs detection. Two stages need to be verified:
- GotCha is capable to capture miRNA
- RCA reaction is triggered by miRNA

Fig.8
Verification of GotCha 664a-3p. ***p < 0.001: Ctrl w/o miRNA compared to Gotcha 664a-3p. ***p < 0.001: Ctrl w/o phi29 compared to GotCha 664a-3p.

Fig.9
Verification of GotCha 33b-3p. ***p < 0.001: Ctrl w/o miRNA compared to GotCha 33b-3p. ***p < 0.001: Ctrl w/o phi29 compared to GotCha 33b-3p.
As shown in Figure 8 and Figure 9, in the presence of miR-664a-3p and miR-33b-3p, there was obvious fluorescence, indicating long ssDNA product was produced. Whereas, there was no obvious fluorescence signal in two control groups. It shows that either containing all component but phi-29 or miR-21 was absence, RCA reaction could not proceed. Therefore, RCA reaction could be performed successfully as designed.
Future Work
Nonetheless, our experiments done so far in the course of the competition did verify various aspects of our kit to a certain extent.
As mentioned above, due to the pandemic, we were unable to complete a fair share of our planned experiments. In the future when restrictions are loosened, we hope to do tests on human blood sample serums, as well as take into account more variables in our experiment (eg. temperature, exonuclease activity in phi29 polymerase) to further validate our kit.

