

ENGINEERING SUCCESS
Circular Probe Design
RCA Modeling
Engineering Success
Circular Probe Design
Pre-test
The circular probe sequence plays an important role in our projects, which is related to whether our kit can capture miRNA and conduct RCA reaction. Initially, we were planning to use existing circular probe sequences for our experiment, only changing the miRNA binding site. However, solely changing of miRNA binding site caused many loops on the predicted DNA structures in this scenario, making the DNA sequence unsuitable for use as the loops may intefere with amplification by polymerase.
The table below presents the prediction of secondary structure while only changing miR binding site. Evidently, only changing the miRNA binging site cannot form a suitable circular probe.

Design And Build
Through consultations with esteemed professors such as Dr. Yao (for more information, see Human Practices), they give us two advices:
- We should design the whole sequence of circular probe including functionless sequence to avoid extensive loops formation and binding.
- We should analyze the secondary structure and thermodynamics of DNA sequences through modeling websites.
Therefore, we designed our own circular DNA sequence a model: Circular DNA Sequence design, which can be found on Github here. This model can effectively avoid the formation of significant loops and ensure thermostability of DNA circular probe sequences. The model follows two principles:
- Avoid complementary base pairs. If there are three or more complementary base pairs in a single DNA strand, it is highly possible forming stem-loops.
- Design non-complementary regions.Non-complementary sequence can prevent error binding of miRNA and immobilisation probe, ensuring complete assembly of GotCha.
Test
We used three DNA secondary structure prediction websites RNAstructure, UNAfold and Kinefold to evaluate the secondary structure and thermodynamic stability of our circular DNA sequences.
From the results, it shows that the DNA sequences generated by our code possessed high thermostability without significant loop formations in our environment. This validates our code.

Learn
Through this process, we learned how to use code to avoid the formation of significant loops and increase stability of DNA (Model). Through ensuring the validity of our code through websites, we have also learned that this process of predicting DNA structure through coding is a complicated one. Therefore, in the future, we hope to dive more into including thermodynamics into our DNA structure prediction. Please anticipate our improved version of code in the near future.
RCA Modeling
Design
Due to the restrictions imposed by the Covid-19 pandemic, we were unable to complete our lab experiments. Therefore, we decided to design a system of RCA modeling for DNA product. Through extensive research, we derived that the mechanism of phi29 polymerase can be modeled by the Michaelis-Menten kinetics equation[1][2]. By fine-tuning the mathematical model to fit our RCA mechanism and environment, we proposed a set of modeling to overcome the obstacle of the lack of experiment data.
Build
As mentioned above, we used the Michaelis-Menten equation as a basis of our phi29 kinetics modeling.
The following is the original mode of action posited by the Michaelis-Menten Equation:

The following is the updated mode of action we employed to suit our experiment:

The main reason why we chose to introduce this change to the Michaelis-Menten mode of action is the mechanism of the phi29 polymerase. It is known through research that the phi29 polymerase elongates primers only when substrate dNTP is attached to the enzyme[3].
The below equation was then derived from the mode of action established.
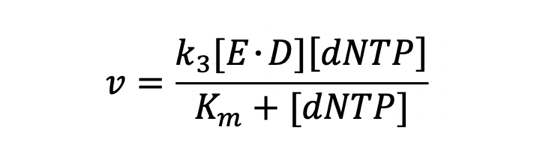

For more information on definitions of variables used, please refer to Model.
The above equations allow us to derive the total amount of DNA product from our mechanism. We then alter the concentration of dNTP and phi29 polymerase to establish a relationship of the variables with the efficiency of RCA mechanism.
Test
We use the relationship graph of fluorscence intensity and DNA concentration derived from our experiments to verify whether the model we designed is corresponed to the result of RCA experiments. The RCA model designed by Dry lab simulates the reaction environment, predicting DNA product under different scales of microRNA concentration .Besides, we found the relationship of fluorescence intensity and DNA concentration derived from Evagreen dye experiments through research. Therefore, we can also derive a relationship graph which shows DNA product under different scales of microRNA concentration. The table below presents the comparison between the prediction of RCA model and experiment result.

Learn + Improve
On the table above we can find there is a clear difference. Through data research, we found that there are undoubtly still more factors ought to be taken into consideration to establish a holistic model for RCA mechanisms. For instance, metal ion and exonuclease activity of phi29 polymerase have certain influence on RCA reaction. Though we were unable to conduct further verifications in the above factors to justify the differences, we still fulfill the improved models. Metal ion influence: According to Robert J. Bauer et al.[4], metal ions are also a major factor affecting the action of polymerase. Km, K3 are the Michaelis constants, and [M2+](Mg2+ or Mn2+) is the concentration of the divalent cation included in the reaction. Data obtained from M2+ titration reactions were fit to a Michaelis-Menten equation with a cooperativity (n) parameter. For those reactions where inhibition was observed at higher concentrations of M2+, an inhibition reaction equation was used instead:

Exonuclease activity: According to Cristina Garmendia's research[5], exonuclease activity of phi29 polymerase will certainly affect the reaction rate of phi29 polymerase.

They derived the rate formula affected by exonuclease activity through the equation above.


In these formulae, Di and (Di•N) represent the concentration of a certain DNA intermediate, i-bases long, and the complex i-mer DNA primer with the next nucleotide to be incorporated bound in its position, not yet catalyzed; both Di and (Di•N) stand for the corresponding complexes with DNA polymerase; Dt is the total amount of DNA polymerase/DNA complexes; N stands for dNTP concentration, and Km = (k-1+ kcat)/k1. For more details, please see Cristina Garmendia's research[5].
Though both of them could be modeled, several parameters in the equations could only derived from experiments. In the future to accomplish our work, we plan to design and conduct experiment incorporating metal ion concentrations and exonuclease activity of phi29 polymerase.
References
- Mohsen MG, Kool ET. The Discovery of Rolling Circle Amplification and Rolling Circle Transcription. Acc Chem Res. 2016 Nov 15;49(11):2540-2550. doi: 10.1021/acs.accounts.6b00417. Epub 2016 Oct 24.
- Morin JA, Cao FJ, Lázaro JM, Arias-Gonzalez JR, Valpuesta JM, Carrascosa JL, Salas M, Ibarra B. Mechano-chemical kinetics of DNA replication: identification of the translocation step of a replicative DNA polymerase. Nucleic Acids Res. 2015 Apr 20;43(7):3643-52. doi: 10.1093/nar/gkv204. Epub 2015 Mar 23.
- Saturno J, Blanco L, Salas M, Esteban JA. A novel kinetic analysis to calculate nucleotide affinity of proofreading DNA polymerases. Application to phi 29 DNA polymerase fidelity mutants. J Biol Chem. 1995 Dec 29;270(52):31235-43. doi: 10.1074/jbc.270.52.31235. PMID: 8537389.
- C Garmendia.et al.The bacteriophage phi 29 DNA polymerase, a proofreading enzyme.J Biol Chem. 1992;267(4):2594-9.
- Robert J. Bauer, Michael T. Begley, and Michael A. Trakselis,Kinetics and Fidelity of Polymerization by DNA Polymerase III from Sulfolobus solfataricus,Biochemistry. 2012; 51(9): 1996–2007.



