

CONTRIBUTION
Contribution
Circular DNA Design Software
Introduction
Due to the restrictions unfortunately imposed by the Covid-19 pandemic, we were unable to plan and execute any Biobrick contributions through lab work of any sorts. Nonetheless, we still want to make contributions to the field of synthetic biology. To do so, we have decided to do so through building software Circular Probe Design, which will provide much convenience for future iGEMers that choose to adopt similar systems for diagnostic tests.
Function Description
Our Software, built through the Python coding language, is universal for all microRNA sequences to be detected, and hence was also employed in part generation of our circular probes for microRNAs 664a-3p and 33b-3p (for more information on part design, please see Design.
Our design includes two binding sites: probe binding site, miR binding site.
Our software is designed for our kit, which through avoiding stem-loop formation allows for smooth-running of RCA mechanism.
Our interface allows the user to insert the sequence of the target miR, and length of functionless sequence.
After which, using our default immobilisation probe sequence, our software will generate a txt. file in the user's computer with all the sequences that the user can use as their circular probe.
The input interface of our software is shown in the figure below
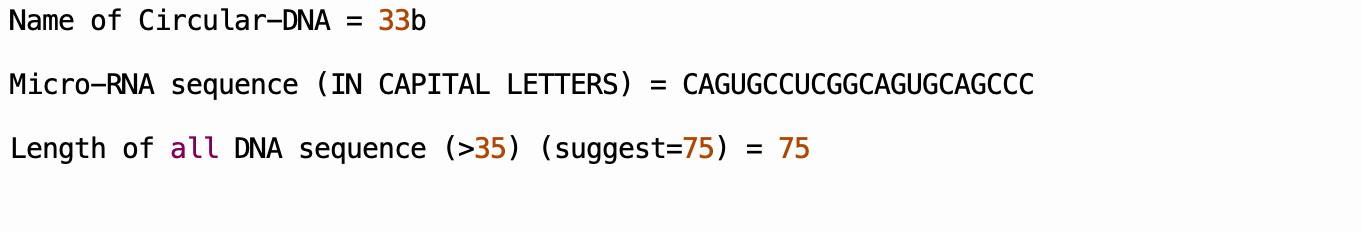
The output interface will display each part of the sequence, as well as the total number of the final sequence and the file name of the output .txt file

Future Work
As the rolling circle amplification mechanism is growing in popularity in the field of synthetic biology and diagnostics, we see the great potential our software can bring to the table in these relevant fields(contribution). In the future, we aim to continue improving our model towards an even more user-friendly software. We recognise that albeit the cut in time our model brings to the process of circular DNA sequence generation, it still lacks the built-in ability to predict DNA structures and value of free-energy, therefore troubling users to manually take an extra step in checking through another website. Therefore, we hope that in the future, this modeling system can be further built on to increase user convenience and provide an efficient platform for both aspiring students such as fellow iGEMers as well as professionals in the field in DNA sequence synthesis.
Experiment Improvement
Overview
Due to the Covid-19 pandemic this year, we were unable to complete all of our experiments as planned. However, for the experiments we managed to conduct, we have listed out the potential problems in handling GotCha for future iGEMers that might want to build on our project.
Variable testing
Firstly, we were unable to complete testing of all the variables that may affect the function, efficiency, and accuracy of our test kit GotCha. If future iGEMers want to take inspiration from our test kit to any extent, it is advicable that they do variable testings to evaluate GotCha holistically.
Temperature Variable
We strongly advice future iGEMers that want to do something similar to our project mechanism to do temperature variable testing in particular. This year, we carried out our RCA mechanism at room temperature, which is 26 degrees Celcius, and this has reaped positive looking results (for more information, see Proof of Concept). Despite this, as the Phi29 polymerase is an enzyme, it is ideal to do temperature variable testing to find out how polymerase activity varies over different temperature, and its optimal temperature of activity to maximise its efficiency and enhance the test kit. For more information, see Protocol.
Serum testing
After finishing the test in the lab, we will conduct the next tests using serum form patients' specimen. In order to verify the specificity and sensitivity of our kit, we will seperate patients into different groups according to their age and cancer stage, and use GotCha to examine their blood serum samples.
Operational recommendations
When handling the magnetic beads, we were met with the problem of beads adhering to plastic surfaces, such as the Eppendorf interior. Therefore, we had to increase the durations for centrifuge and magnetic separation to ensure the complete separation of beads.

