Design
Project design
In our project this year, we have achieved some major criteria that had been chosen to ensure maximum efficacy and safety of our proposed new therapeutic vaccine for TNBC. Our design included three main aspects which are the vaccine design itself, the vector used to deliver it to the host cell, and finally the design of directed evolution algorithms whose main function was to improve the yield of the vaccine and ensure its structural stability and include the ability to increase or decrease the vaccine’s expression depending on the surrounding microenvironment, which also allows our model to be system-sensitive and specific.
As our work is a continuation of last year’s project, we had to improve our previous approach regarding the usage of a multi-epitope DNA vaccine. We did so by adopting another approach via the utilization of Virus-Like Particles (VLPs).
Non-Structural Proteins for EEV-Alphaviral Vector:
We used four non-structural proteins that together forms the main complex responsible for the synthesis positive-sense viral RNAs, results in the synthesis of both the genomic and subgenomic RNAs, of which the subgenomic RNA is produced in excess of the viral genome which allows the virus to self-replicate into millions of copies of the virus.


Figs.1, 2 show a static images of the final model of each one of the 4 NSPs, each 3D representation includes 2 images of the model, the one on the left represented by color-coded chains, while the one on the right representing the quality of the model at each amino acid of the non-structural protein according to the predicted LDDT-score.

Fig.3 shows the aligned errors in terms revealing the degree of uncertainty in the relative position of the two protein chains of the NSP protein dimer. The plots are shaded according to a scale from blue to red, where the blue color represents a least error.
RBP-Riboswitch design:
We introduced the idea of RNA-based regulatory parts, especially the riboswitches as a mean for post-transcriptional control for our synthetic regulatory circuit of the vaccine. The use of Ribosomal binding proteins (RBP) including L7Ae and MS2 in our circuit ensured the applicability and prospect for post-transcriptional regulation of the vaccine circuit. Those regulatory RBPs were selected due to their high specificity of binding to their specific RNA motifs. We characterized and optimized the structure of the riboswitches comprising the (L7Ae & MS2) RBP and their binding motifs (Kink-loops motif (K-turns) and U2-small nuclear ribonucleoprotein motif (snRNP)). Our composite consisted of 2 main RBP-based repressors:
1-L7Ae: K-turn system: This system was modified by linking the L7Ae to an intracellular antibody and dcas13 enzyme representing an L7Ae classifier of the tumour environment. L7Ae (which inhibits the circuit via binding to its k-turns) is consumed once the intracellular antibody or dcas13 recognize and bind to their target mRNA, thus L7Ae inhibitory effect is removed which leads to the expression of the output protein.

Fig.4 shows: Structural characterization of the main free energy (MFE) secondary structure of L7Ae-Kink-turns riboswitch is 25.99 %.
2-MS2:U2-snRNP system: is another RNA-based OFF-switch improved by fusing the MS2 protein-based riboswitch to a destabilizing domain (DD) that is stabilized in the presence of the small molecule trimethoprim (TMP). TMP regulates the inhibitory effect of the MS2:U2-snRNP-riboswitch by binding to the DD-MS2, therefore enabling and enhancing the dimerization of their splitting, finally terminating its inhibitory function.

Fig.5 shows: Structural characterization of the main free energy (MFE) secondary structure of MS2-U2SNRP riboswitch is 35.78 %.
Design of Virus-Like Particles (VLPs):
VLPs are a class of diverse nanoparticles that are formed by structural viral proteins, such as capsids, and can self-assemble. In spite of their resemblance to viruses, they are non-infectious because they lack viral genetic material. Humans respond to VLPs by displaying a high density of antigenic epitopes that mimic the 3D conformation of native viruses and thereby evoke the desired humoral and cellular responses. HBV's core antigen (HBc) has been extensively used for VLP applications throughout the years. The core antigen of HBV is a protein of 21 kDa that self-assembles to package the viral polymerase and pregenomic RNA during HBV replication by the nucleocapsid particles. It shows ease of recombinant expression and self-assembly. Also, it has several characteristics that make it a desirable carrier for foreign haptens. Simply HBcAg acts as a carrier by inserting its peptidic epitopes at different positions. Using this method, foreign haptens will take up the viral neutralizing antibodies binding sites, which will result in reduced HBcAg antigenicity and immunogenicity, and enhanced foreign epitope antigenicity and immunogenicity. 33 years after the first vaccine using this technology was approved by the FDA, vaccines based on virus-like particles have demonstrated their effectiveness in human health.
In Our VLPs, we loaded TNBC epitopes, which we used in last year’s project, onto the Hepatitis B core protein. Then, we tested the results’ functional stability and molecular dynamics and made comparisons between them depending of the oligomeric structure of the VLP, their sequence coverage, predicted IDDT per position, RMSD, RMSF and predicted alignment error.



Figs.6, 7, 8 illustrate our 6 VLPs. The first row in each figure illustrates the oligomeric structure of our VLPs after loading the epitopes on the HBc. The second row shows the sequence coverage and predicted IDDT per position for each VLP. The third and fourth rows show the RMSD and RMSF which calculate the stability and the flexibility of the VLPs respectively. The fifth and final row show the predicted alignment errors.
Vector development:
For the assembly and loading of our circuit, our team chose 2 plasmids as backbone; EEV and pcDNA which allows comparison between alpha virus vector and traditional vectors.
Parts assembly:
The team used Ecor1 restriction enzyme for both pcDNA and EEV plasmids.
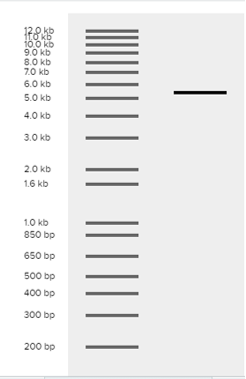
Fig.9 represents the virtual digest using Ecor1 for pcDNA plasmid.

Fig.10 represents the virtual digest using Ecor1 for EEV plasmid.
The vaccine assembly consists mainly of 3 parts (DD-MS2, snRNP-cas12g-L7Ae, and hBax-VLPvacc) that’s why we assembled using 2 plasmids; alpha virus plasmid (EEV) and traditional plasmid (pcDNA).
DD-MS2
For assembly of u2snRNP-cas12g-l7Ae, our team used 2 plasmids as backbone (EEV and pcDNA) which allows comparison between alpha virus vectors and traditional vectors. Another designed system includes a different riboswitch which consists of the MS2 protein that binds with small nuclear ribonulceoprotein U2-snRNP to inhibit transcription. In our design MS2 is fused to destabilizing domain (DD) by Gly-Ser linker which could be stabilized after administering Trimethoprim (TMP). TMP which acts as a small molecule inhibitor will free the MS2 after stabilizing DD. Free MS2 binds to U2-snRNP which inhibit the circuit so as to be able to control the transcription process in extreme conditions as cytokine storm or unpredictable results.
Golden gate:
pcDNA
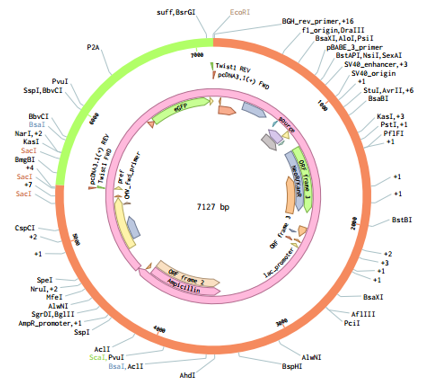
Fig.11 Assembly of MS2-DD (pcDNA), Golden gate.
EEV

Fig.12 Assembly of MS2-DD (EEV), Golden gate.
U2snRNP-cas12g-l7Ae
For assembly of u2snRNP-cas12g-l7Ae, our team used 2 plasmids as backbone (EEV and pcDNA) which allows comparison between alpha virus vectors and traditional vectors.
Cas12g is used in conjugation with L7Ae protein (that has an inhibitory effect on the transcription by binding to its kink-turn or snRNP) by Gly Ser linker. The system simply recognizes and binds to mRNA in the cancerous environment which leads to consumption of L7Ae protein. Therefore, not binding to kink-turn or snRNP to inhibit the inhibitory effect on the transcription which finally leads to stimulation of the transcription in order to increase the yield of the vaccine. That is why, it is a cell specific design by binding to mRNA of PD-L1 which has an immune evasion role in the cancerous environment especially TLCs.
Golden gate
pcDNA

Fig.13 assembly of u2snrp-cas12g-l7ae (pcDNA plasmid).
EEV

Fig.14 assembly of u2snrp-cas12g-l7ae (Alpha virus vector).
hBAX-VLP assembly
For hBAX-VLP assembly, our team used 3 plasmid as backbone (EEV and pcDNA) which allows comparison between alpha virus vectors and traditional vectors.
Our TNBC vaccine is supposed to be injected intra-tumorally. That is why an oncolytic activity is added to the circuit via hBax to perform an apoptotic function which helps in both combating cancer cells and apoptotic bodies which release neo-epitopes to be recognized by immune cells.
Golden gate
pcDNA

Fig.15 Assembly of hBAX-VLP (pcDNA plasmid), Golden gate.
EEV

Fig.16 Assembly of hBAX-VLP (EEV plasmid), Golden gate.
Alphaviral_FADD_L7Ae_Hbax_VLP assembly:

Fig.17 Assembly of alphaviral_FADD_L7Ae_Hbax_VLP using Digest and ligate method.

Fig.18 Assembly alphaviral_FADD_L7Ae_Hbax_VLP using Gibson method.

Fig.19 Assembly alphaviral_FADD_L7Ae_Hbax_VLP using Golden Gate method.

