PROOF OF CONCEPT
Fluorescence Limit of Detection
In order to test the device’s ability to detect fluorescence, characterization experiments were conducted with the fluorescent dye, Lucifer Yellow, so that the concentration of dye and thus the expected relative fluorescence would be a known variable. Starting with the stock solution of the dye, serial dilutions of the dye by differing factors were created to determine lower and upper limits of detection as shown in the figure below.

Optical Density Calibration
The device’s ability to read the optical density of a cell culture accurately and precisely was tested in a very similar way to how the fluorescence detection was tested. E. coli. were grown to a high density culture and then serially diluted using Luria Broth media. The actual ODs of the bacteria were measured with a Tecan, a standard state-of-the-art plate reader. The readings correlated to the concentration of the bacteria in a sample. Each dilution was then plated in every well of the plate and measurements were taken with our engineered device. Once again, 100 readings per well were taken and averaged so that the output from the experiment was one averaged reading per well for a given dilution and the OptoReader optical density measurements were correlated to the measurements obtained with the Tekan.
To find the optimal intensity of all OD LEDs, the OD of several dilutions of cell culture was measured with the intensity of all OD LEDs varying on a scale of 200-800 (the total range of the intensity of the LED runs from 0-4095 in arbitrary units). Based on the experiments, it was found that intensity of 500 gave the most linear relationship of OD readings with the OptoReader compared to the industry standard plate readers. We noted that the signal from the OD LED varied greatly from well to well, likely due to the fact that OD measurements are more sensitive to the angle of light exposure. To calibrate OD readings, we manually adjusted the intensities of each OD LED so that their signal was more uniform relative to each other when read by their respective photodiodes.


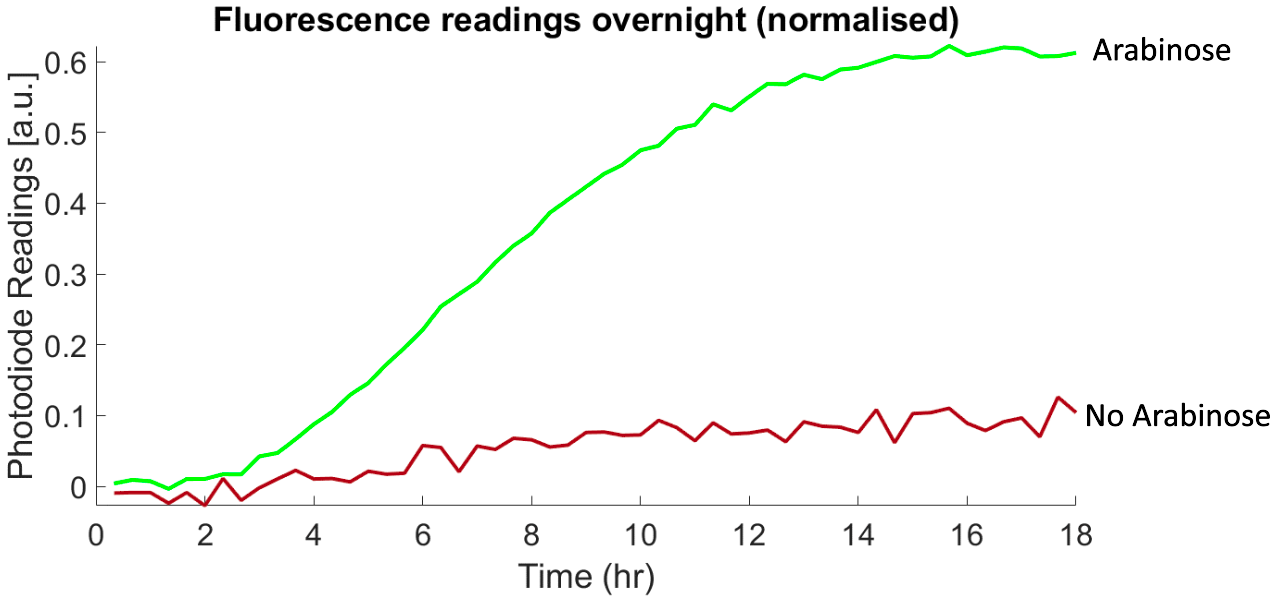
Overnight in Bacteria
In order to test the suitability of the device to conduct experiments over long time scales, a series of experiments were conducted over a period of 18 hours. We collected both optical density and fluorescence measurements of E. coli that could express the fluorescent protein mAmetrine upon induction with arabinose. Over 18 hours, we confirmed that bacteria could thrive in the device, and the device could successfully measure growth (OD) and fluorescence increases after induction.

Stimulating an Opto-Tool
After validating that each part of the OptoReader worked independently (LEDs and photodiodes), we aimed to test our device in a biological system. Using the pDusk system, we were able to show light-dependent protein production in E. coli.
pDusk is a system that activates transcription of a fluorescent protein (we cloned mAmetrine) in darkness and represses gene expression in light
Over 18 hours, we tested two light conditions: darkness and 10% light dosage. Results showed a significant difference in mAmetrine production in illuminated vs. unilluminated wells as indicated by fluorescence values normalized by optical density measurements. Thus, the OptoReader has proven to be able to monitor cell growth, read fluorescent output, and successfully activate/deactivate opto-tools via light-controlled gene expression taking high-throughput data at consistent reading intervals that could not have been achieved through manual reading methodologies that were previously available.


Feedback Control
In this experiment, we implemented the feedback function in our software to show that feedback no only is possible in the hardware of the device, but also leads to biologically relevant data as well. We programmed our feedback algorithm to stimulate cells with blue light initially, but to turn off light stimulation once the cell density in the well reached an OD of 0.4 and greater. To do so, the OptoReader monitored optical density by taking readings every 30 minutes. Once the cells grew to an OD of 0.4, the light stimulation was permanently turned off. We tested this in E. coli transformed with pDawn system which harnesses the lac operon to activate transcription of a target gene (fluorescent protein mAmetrine in our case) in light. We see activation of mAmetrine transcription in pDawn while cells are stimulated with blue light. At hour 10.5, the cells reached an optical density of 0.4 and we observed inflection points in the fluorescence curve at this time. While the changes in fluorescence at hour 10.5 are slight, this may be due to the fact that the bacteria are nearing saturation in growth, altering transcriptional dynamics. Future experiments will repeat this test using a lower OD to trigger feedback. With feedback mechanisms like this, we are able to study and modulate optogenetic tools in ways that have previously been impossible, opening new avenues of study.










