Overview
With climate change, temperatures throughout the world are becoming more extreme and unpredictable, adversely impacting entire ecosystems. At a smaller scale, farming is particularly affected by these drastic weather fluctuations. For instance, in the Swiss region of Valais, we have observed repetitive freezing spells in the past few springs, causing devastating losses in crops, such as apricot trees. In order to solve this problem, we developed three complementary approaches and applied the engineering cycle (Fig. 1).

Antifreeze Proteins
To reduce frost damage caused by ice crystals that destroy sensitive plant tissues during late spring freeze, we wanted to develop a solution containing antifreeze proteins (AFP). These proteins bind to ice crystals and thereby inhibit their growth. We did extensive research to learn more about how AFPs work and found out that they naturally exist in organisms living in cold temperatures. The proteins allow them to be resistant to freezing. Their activity can be characterized using either thermal hysteresis (TH) or ice recrystallization inhibition (IRI). TH describes the difference between the non-equilibrium freezing and melting temperatures [1]. IRI activity prevents the growth of larger ice crystals, which could damage plant tissues, at the expense of smaller ones [2]. Not all AFPs have the same activity. For this project, we therefore chose three antifreeze proteins, each one with a different activity. This allowed us to make comparisons and combine different AFPs to develop a solution efficient for our purpose. We chose to work with antifreeze proteins from Rhagium inquisitor (RiAFP), Daucus carota (DcAFP), and Flavobacterium frigoris PS1 (FfIBP).
First, we designed 6 new composite parts by inserting the three different AFPs into two vectors, pET-17b and pCold-I. The pET-17b plasmid contains a T7 promoter for regulated high-level expression of recombinant proteins. The pCold-I plasmid contains a cold-shock protein A (cspA) promoter for expression of proteins that are more difficult and cannot be expressed with the T7 system. We therefore compared expression of AFPs using each vector with the aim to optimize expression and synthesis of antifreeze proteins.

The various constructs (Fig. 2) were built and transformed into Escherichia coli BL21 (DE3) cells. We chose to work with E. coli BL21 (DE3) because it allows for high level expression and since the aim of our project is to develop a spray for apricot crops, large scale protein production would be necessary. E. coli is a commonly used organism and therefore easily available.
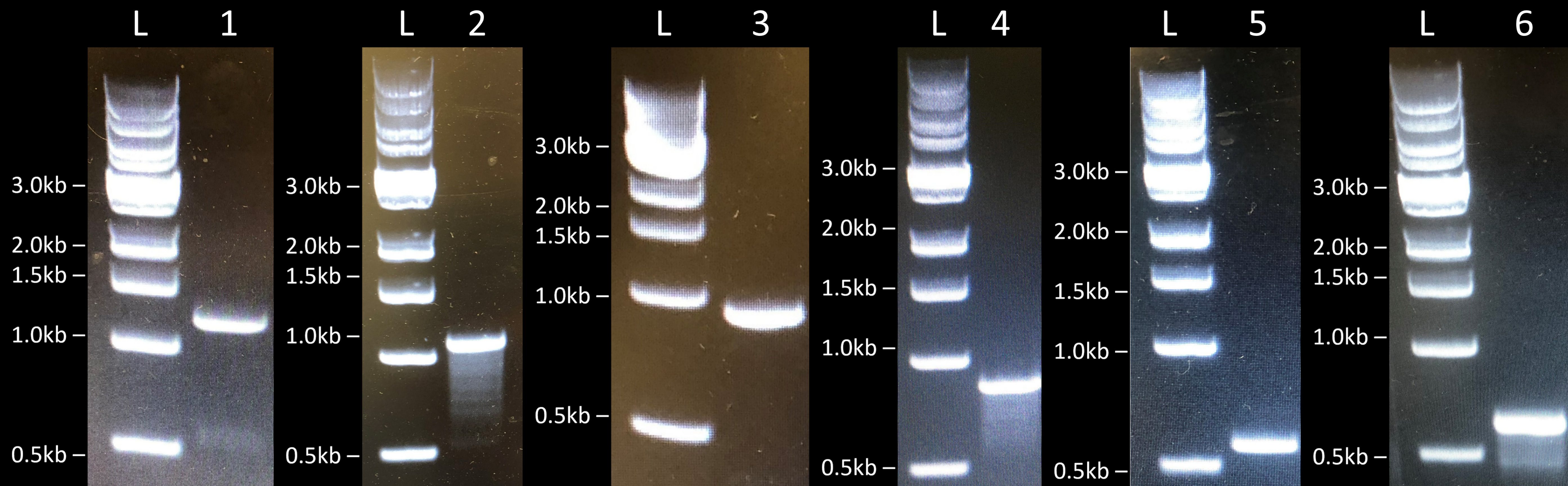
Initially, we first amplified the various AFP genes by PCR and linearized the plasmids by restriction enzyme digestion. Then, we assembled the various constructs by Gibson Assembly, transformed E. coli BL21 (DE3) cells by electroporation, and performed colony PCR to check if the genes were correctly assembled in the various constructs (Fig. 3). We obtained the expected band sizes and confirmed their sequence by sanger sequencing. We therefore concluded that we managed to successfully clone FfIBP, DcAFP, and RiAFP in E. coli BL21 (DE3) cells.
Next, the various AFPs were expressed in E. coli BL21 (DE3) using IPTG and then purified by His-tag affinity column and gel filtration. To test if the expression worked, we ran total cell SDS-PAGE (15%) of the different E. coli BL21 (DE3) cultures stained with Coomassie Blue.
We used two different vectors, pET-17b and pCold-I, to compare how the promoters influenced expression efficiency for both AFPs. The expression was successful for FfIBP and RiAFP in both pET-17b and pCold-I plasmids. We observed thicker bands that matched the weights of the different antifreeze proteins (FfIBP 28.4kDa, RiAFP 12.8 kDa). The pCold-I vector worked better for both RiAFP and FfIBP, and the bands on the gels appeared to be thicker for this plasmid (Fig. 4 and Fig. 5). This can be explained by the fact that the natural expression of AFPs happens at cold temperatures, and any expression system adapted to these conditions will be more efficient. We induced expression at 15°C. At this temperature the 5’UTR of the mRNA from the pCold-I plasmid adopts a highly stable secondary structure and the TEE sequence then improves translation initiation of our AFP genes through a ribosome trapping process. The pET-17b plasmid does not have these features and is therefore less efficient. We can conclude that the pCold-I plasmid is a better system for expression of cold-stable proteins like AFPs.


Once expression was successfully induced, we purified the AFPs and ran an SDS-PAGE (15%) of the purified recombinant proteins stained with Coomassie Blue. More information on this can be found here.
The final test was the functionality of the proteins, which we did using two different assays. We measured the TH activity of the purified recombinant proteins and evaluated their effect on frost damage on plants. The details on these measurements can be found here.
Tailocins
Our main goal was to extract tailocins, phages-derived bacteriocins, to eliminate a bacterium present on apricot tree, P. syringae pv. syringae. This pathogen produces ice nucleation proteins to catalyze the formation of ice and therefore damage the plants’ membrane. Since we were not able to obtain the exact strains found on apricot trees, we therefore chose a closely related strain, Pseudomonas syringae pv. syringae B301D, as our target strain, since its genome sequence was available and this strain’s sensitivity to other Pseudomonas tailocins had already been characterized by Baltrus et al.[3]. Our strategy was therefore to produce tailocins from the strain Pseudomonas syringae pv. aptata DSM50252 which were shown to target Pseudomonas syringae pv. syringae B301D and proved that the produced tailocins have the ability to kill our target strains. To extract tailocins, we based our protocol on Baltrus et al.'s one with modifications suggested by Jordan Vacheron, from the University of Lausanne[3]. Briefly, we exposed our producing bacteria to mitomycin C, which triggered tailocins expression and cell lysis. We then purified tailocins from the lysis using PEG precipitation. Next, we tested their efficiency using an overlay essay. This method consists of serially diluting the purified tailocin solution and spotting the dilutions on a soft agar plate inoculated with our target strain. With this assay, we have successfully proven that our target strain lyses in the presence of our two purified tailocins solutions.

Furthermore, we have proven that killing our target strain significantly reduces the freezing point of water. We compared two drops, one with our target strain and the tailocins and the other only with the target strain. Then we compared the freezing time using FROZONE.The results prove that killing Pseudomonas syringae pv. syringae B301D, reduce the freezing time significantly.

Having confirmed the efficiency of our tailocins approach in reducing Pseudomonas syringae pv. syringae B301D induced ice nucleation, we aimed at cloning the tailocins gene cluster in E. coli.
Phages
The goal of this part of the project was to delete InaZ, the gene responsible for the production of Ice Nucleation Proteins (INP). INPs are harmful to fruit trees [4], since they allow the formation of ice crystals at a higher temperature compared to INP-free water, causing damage to plant epidermal cells, and thus disabling some of its physiological functions such as flowering [5]. So, by deleting the initial source of these proteins, in our case InaZ, we aim to disarm P. syringae pv. syringae, rendering it harmless to the plant. To achieve the deletion of this gene, we want to make use of the homologous recombination (HR) mechanism in the strain. The reason why we chose this repair mechanism is because the non-homologous end-joining (NHEJ) one appeared to be absent in this bacterial strain, contrarily to HR dedicated proteins [6]. To increase our chances to make the recombination happen, we decided to induce a DNA damage [7], notably a double-stranded break, into InaZ coding sequence.
The engineering procedure required the construction of a phagemid (i.e. a viral-derived plasmid) [8], the carrier of the two biological elements necessary for our goal. These two parts are a single-guide RNA (sgRNA), associated with the CRISPR/Cas9 cassette, which allows the DNA cutting machinery Cas9 to be directed towards a specific DNA sequence in the INP coding sequence. Three different sgRNAs were designed to target three different regions of InaZ, allowing us to select the most efficient in targeting and cutting this gene, but also the best to increase the chances of homologous recombination occurring.
The second element required for the construction of our engineered phagemid is called “Donor Sequence” and consists of a 2 kb sequence consisting of two regions homologous to the upstream and downstream sequences of InaZ. This double-stranded sequence provides the perfect template for the homologous recombination to occur. The InaZ coding sequence will eventually be deleted and replaced by the donor sequence in the P. syringae pv. syringae’s chromosome, as a result of the HR mechanism [9].
A schematic overview of this mechanism is represented here:

To trigger the mechanism shown in figure 8, the engineered phagemid needs to be efficiently delivered into P. syringae pv. syringae. This is provided by using a defective P1 phage that can carry our phagemid [10]. The resulting virions would eventually be harvested and put into our spray product.
The first step is to clone each of the three guide RNAs into a different plasmid unit containing a CRISPR/Cas9 system (pJH1) by using Golden Gate assembly. The insertion is made in the CRISPR sequence between the two BsaI sites [11].
We succeeded and obtained three different plasmids, each of them carrying one of the three sgRNA (Fig. 9).
The Golden Gate Assembly protocols can be found here.



The Gibson Assembly protocols can be found here.

We decided to test the efficacy of our constructs in deleting InaZ. As a proof of Concept, we transformed two different plasmids carrying either sgRNA 1 or 2 and the donor sequence into Pseudomonas syringae pv. syringae B301D.
After transformation, the bacteria were plated into LB agar plates supplied with Kanamycin 50. The next day, we picked the colonies and checked the clones by performing colony PCRs to confirm the deletion of InaZ. Unfortunately, all grown colonies still carried InaZ in their genome, as shown in figure 11. The expected PCR product confirming InaZ deletion would have been around 2 kb, whereas a PCR product of about 6 kb would mean that InaZ is not deleted.

There can be several things that might not have worked. First, we might need to test if Cas9 got expressed. We could perform a Western Blot to test Cas9 expression. Second, we could test the efficiency of the gRNAs. We could transform another plasmid containing the target region of the gRNAs and see if Cas9 induces a cut in this plasmid in P. syringae. Third, homologous recombination might have been low efficient in P. syringae, especially when the designed homology arms are not adjacent to the cut site. We could design a donor sequence carrying homologies adjacent to the cut site, which will not completely delete InaZ but at least disrupt it. Fourth, instead of one cut, we could cut InaZ twice by using two gRNAs targeting the start and end of the gene. This might facilitate the integration of the donor sequence by the HR repair mechanism, as the sequences next to the cut are directly homologous to the ones provided by the donor sequence. With these measures, we might be able to get our strategy work. In parallel, we tested two more strategies that look more promising.
Hardware and Software
To perform some of our experiments, a precise control of the temperature was needed. We therefore created FROZONE, a precise cooling device that fits under a microscope, acting similarly to a Nanoliter Osmometer. The design of our machine was inspired by the MicroIce LTD. According to MicroIce, "A Nanoliter osmometer is a cooling stage mounted on an upright optical microscope. Cooling of the stage is achieved with the use of Peltier devices driven with a precision temperature controller."
Our nanoliter osmometer, FROZONE, was designed and created by us. The cooling of the device is achieved with a thermoelectric cooler controlled by a custom Python software. We also designed a vacuum chamber to minimize the crystallization of ambient humidity on our samples. Put simply, we created a machine that precisely controls the temperature of a copper plate in order to measure various aspects of the solutions we produced.
The build of our machine was not successful on the first try - we had to make, test and improve many prototypes before being satisfied with our final product.
For more information about software click here.
Vacuum chamber
When we first designed our machine, we did not think to add a vacuum chamber around the copper plate on which we froze the samples. However, with this setup, we observed lots of undesired droplets forming on the plate due to the ambient humidity (Fig. 12).



We then realized that creating the vacuum with our sample drop already on the plate would often lead to the evaporation of the drop (Fig. 13). We therefore adjusted our protocol to create the vacuum just before adding the sample.
To absorb any moisture residue present in the vacuum surrounding our sample, we also added some silica beads inside our vacuum chamber. With this setup, we were able to capture the freezing of our sample without interference from ambient humidity or the evaporation of our solutions. (Fig. 14).
Top glass
We initially placed a single layer of glass over the copper plate to create the vacuum chamber, but condensation had a tendency to form on the outside of the glass. We therefore modified this glass to make it double layered, successfully preventing the formation of condensation. Between the two layers of glass, we placed silica gel beads to absorb any moisture.
Light
The microscope we used to visualize the freezing of our solution drops on FROZONE’s copper plate had no top light, so we initially used a lamp to illuminate the machine. However, this LED lamp caused the heating of the plate, modifying this essential parameter. It also did not generate enough light, making it impossible for us to zoom in the images we captured. We instead decided to use an optical fiber light source to light up our copper plate. This light source was much more focused, allowing the proper illumination on our samples. This kind of lamp does not generate heat, so its use did not affect the temperature of our machine.
Software - temperature control
At the beginning, we controlled the temperature on FROZONE through a graphical interface, on which we had added a slider - we needed to scroll this slider up and down to modify the temperature.
This was not the most efficient way to set the temperature on our machine, so we decided to replace the slider with an input box. This allowed us to enter the desired temperature with our keyboard. This mechanism worked well to set a temperature, but was limited in the fact that it was more difficult to gradually increase or diminish the temperature. We therefore designed a spinbox, allowing us to raise or diminish the temperature on our machine using the up and down arrows on our keyboard. Each time we press an upward or downward arrow on our keyboard, we can now raise or diminish the temperature of 0.1 °C respectively.
For more information about software click here.
References
- [1] A. K. Gruneberg et al., Ice recrystallization inhibition activity varies with ice-binding protein type and does not correlate with thermal hysteresis, In: Cryobiology, Vol. 99, pp. 28-39, April 2021 Available from: https://doi.org/10.1016/j.cryobiol.2021.01.017
- [2] M. Chow‐Shi‐Yée et al., Inhibition of ice recrystallization and cryoprotective activity of wheat proteins in liver and pancreatic cells, In: Protein Sci., Vol. 25, No. 5, May 2016 Available from: /pmc/articles/PMC4838640/
- [3] D. A. Baltrus et al.,Localized recombination drives diversification of killing spectra for phage-derived syringacins, In: The ISME Journal, Vol. 13, pp. 237–249, 2019 Available from: https://www.nature.com/articles/s41396-018-0261-3
- [4] S. E. Lindow, The role of bacterial ice nucleation in frost injury to plants, In: Annual Review Phytopathol, Vol. 21, pp. 363-384, 1983 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1065830/
- [5] S. J. Roeters et al., Ice-nucleating proteins are activated by low temperatures to control the structure of interfacial water, In: Nature Communications, Vol. 12, No. 1, pp. 1183-1192, 19 February 2021 Available from: https://www.nature.com/articles/s41467-021-21349-3
- [6] B. Swingle et al., Recombineering Using RecTE from Pseudomonas syringae, In: Applied and Environmental Microbiology, Vol. 76, No. 15, 11 July 2010 Available from: https://journals.asm.org/doi/10.1128/AEM.00911-10
- [7] P. J. G. Rauch et al., The expression of the Acinetobacter calcoaceticus recA gene increases in response to DNA damage independently of RecA and of development of competence for natural transformation, In: Microbiology, Vol. 142, Pt. 4, pp. 1025-1032, 1996 Available from: https://pubmed.ncbi.nlm.nih.gov/8936328/
- [8] H. Qi et al., Phagemid Vectors for Phage Display: Properties, Characteristics and Construction, In: Journal of Molecular Biology, Vol. 417, No. 3, pp. 129-143, 30 March 2012 Available from: https://pubmed.ncbi.nlm.nih.gov/22310045/
- [9] K. Sakaguchi et al., A targeted gene knockout method using a newly constructed temperature-sensitive plasmid mediated homologous recombination in Bifidobacterium longum, In: Applied Microbiology and Biotechnology, Vol. 95, pp. 499-509, 2012 Available from: https://pubmed.ncbi.nlm.nih.gov/22639142/
- [10] J. T. Kittleson et al., Scalable Plasmid Transfer using Engineered P1-based Phagemids, In: ACS Synthetic Biology, Vol. 1, No. 12, pp. 583-589, 21 December 2012 Available from: https://pubs.acs.org/doi/abs/10.1021/sb300054p
- [11] J. Ho et al., The application of the CRISPR/Cas9 system in Pseudomonas syringae pv. actinidiae, In: Journal of Medical Microbiology, Vol. 69, No. 3, pp. 478-486, March 2020 Available from: https://pubmed.ncbi.nlm.nih.gov/31935181/
- Figure 8 created with Biorender.com
