Engineering



Changes to Parts
Part BBa_K3903000 – Non-ribosomal peptide synthetase (NRPS)
We decided to order our genes in the form of gblocks from IDT, and we checked our gene sequences using IDT’s gblocks tool here. All our Shinogen genes were satisfactory for manufacturing, apart from non-ribosomal peptide synthetase (NRPS) which had a section of bases within it that increased its difficulty score above 7. This issue would have made our order hard to synthesize and taken too much time for our laboratory schedule. The manufacturing issue was due to a stretch of DNA sequence having too many guanine bases next to each other (starting at base 1111).
To avoid this, we changed the sequence from GGG GGG GTG GGC G to TTA GGC GGT TGG GCC, which was also codon optimized for the abundance of nucleotides in E. coli. This change was then re-checked with the IDT tool, and the difficulty score had been reduced enough that we were able to use this new sequence in our order. All the genes were then checked to have the ATG start codon (which is needed in E. coli).
This part is an updated CDS of the codon optimised NRPS from our Phase 1 project (Part:BBa_K3634003), which now has a slightly changed sequence to accommodate manufacturer (IDT) specifications. NRPS is the final constitutive gene in the Shinogen cluster which converts sedoheptulose 7-phosphate to shinorine. The product, shinorine, has UV protectant properties as categorised by the Minnesota 2012 iGEM team (BBa_K814003). The gene was taken from cyanobacteria species Anabaena variabilis (ATCC 29413) and optimised for E. coli by our 2020 team. This year, it has been further corrected for IDT manufacturer.
Due to this change, NRPS was able to be manufactured, and therefore we have contributed to the registry with a part that is ‘ready to go’ and can be utilised by future teams looking to conduct research into the Shineogen pathway or other pathways that utilise this gene. Although this part is now fit to use, it could be further improved in the future by the re-addition of the composite part (with ATPG) to create a constitutive gene block with both sections of the gene pathway included (something our 2020 team discuss in silico, but this would need to be achieved in the lab for an improvement). The link to the new part can be found at the link here.

List of Parts
Shinogen
The parts we used this year in the lab:
ATPG
NRPS
DHQS
0-MT
Last year’s team also constructed composite parts for each of our genes (shown below); however, these became redundant in our experimental design as we simplified the construct to allow for IDT manufacture. The composite parts were too complicated to be produced and we did not need the genes to have separate promoters, ribosome binding sites (RBS), or terminators as our chosen plasmids already contained these.
DHQS and O-MT
ATPG and NRPS
Thanogen
The Thanogen parts (BBa_K3634000 to BBa_K3634021), designed by the 2020 team, were going to be ordered if there was time towards the end of our project; however, these parts were not fit for IDT manufacture. They were very complex to adapt to meet the regulations; therefore, time was dedicated to implementing the Shinogen construct and not on rectifying these sequences. Though improving the Thanogen parts to optimize them for manufacture is something that could be done in the future for later testing and development of the gene circuit.
ccaS
ho1
pcyA
ccaS/ho1/pcyA Composite Part
LacO- and LacP
ccdB
Glucose Mediated Death Sensor
ccdAB Promoter/Operator
CcdAB-Controlled mf-Lon Protease
Lac and mf-Lon Degradation Tag
Placlq and Lac Repressor (with mf-Lon Degradation Tag)
ccaR
PL8-UV5
ccaR-Mediated ccdA Expression System
CviJI Endonuclease (with ssrA degradation tag)
LacI-Controlled CviJI Endonuclease (with ssrA degradation tag)
Plasmid and Bacteria Strain
Plasmid
For plasmid design we ended up having to do a significant amount of work to adjust the in-silico design to one that would fit into a laboratory experimental design. After looking through the work that the 2020 team did on the project, we found that their proposed plasmids (pSB3B1-DOPH and pSB3E1-AN) were actually theoretical designs and not existing plasmid vectors. We did try to search those names and came up with no existing plasmid constructs.
 The theoretical plasmid constructs designed by the 2020 team (pSB3E1-AN and pSB3B1-DOPH).
The theoretical plasmid constructs designed by the 2020 team (pSB3E1-AN and pSB3B1-DOPH). Following this, we decided to still follow the general plasmid structure design by last year’s team but just find physical plasmids we could use. With some advice from our lab supervisors Simon and Jacqueline, we found two compatible plasmids to use (pET-DUET-1 and pACYCDuet-1). Each of these pET plasmids had two individual cloning sites with their own promoter, RBS, and terminators which was perfect! These two plasmids were ideal since we could put in more genes and have an easier time doing it, also pETDuet-1 had ampicillin resistance and pACYCDuet-1 had chloramphenicol resistance.

 The two plasmids we found that would suit our project (pACYCDuet-1 and pETDuet-1).
The two plasmids we found that would suit our project (pACYCDuet-1 and pETDuet-1).
These two Duet-1 plasmids we decided on also had protein tags in their cloning sites, and we would use this in the future when we got to protein expression and purification (for easier identificiation).
We did end up changing from pACYCDuet-1 to pRSFDuet-1. Why we made this descision was due to numerous factors. The primary reason was because pRSFDuet-1 had kanamycin resistance, and this made it easier for us as we would need to order in chloramphenicol (we could access pre-poured ampicillin and kanamycin plates from the biology department in St Andrews). Lastly, pACYCDuet-1 and pRSFDuet-1 had the same genetic sequence for their cloning sites, so we didn’t need to change our Gibson assembly primers at all.
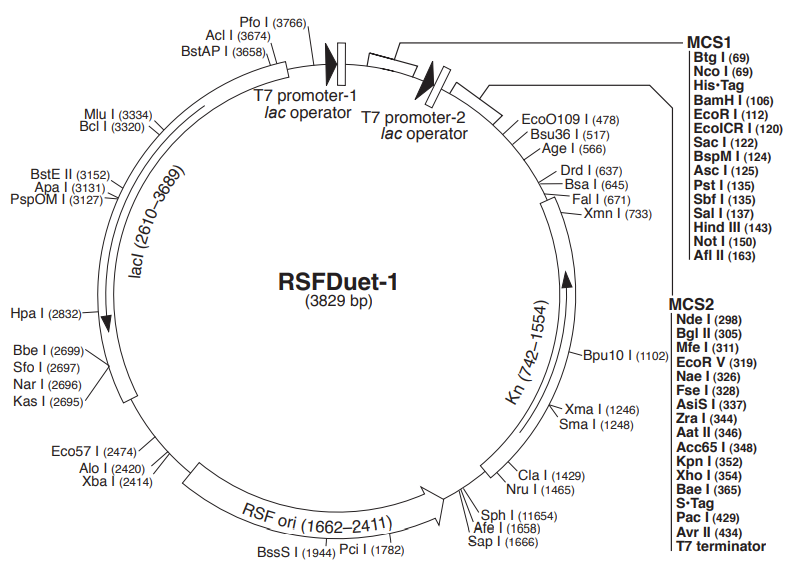 Bacterial Strains for Laboratory Experiments
Bacterial Strains for Laboratory ExperimentsThe wetlab team agreed to use both an expression and cloning strain for our laboratory experiments. The reason we chose an additional cloning strain was because they have higher plasmid replication rates and would be used while we worked on gene insertion. For the cloning bacterial strain, we chose to go with the E. coli strain DH5a since it is classified as Biosafety Level 1 and commercially availiable. For our expression strain we decided to use BL21, which was BSL1 and an E. coli we could access, as we were not able find Nissle E. coli for purchase online.
However, we were able to source our plasmids, DH5a and BL21 E. coli strains from local St Andrews resarchers. We wanted to thank everyone who lent us parts, inlcuding the plasmids, bacterial strains reagents, advice, and overall support!
