
This project used Saccharomyces cerevisiae as the chassis organism to produce a new sunscreen substance called gadusol. In the synthesis pathway of gadusol, S7P of the pentose phosphate pathway is an essential substrate. According to the literature review, introducing the xylose utilization pathway in Saccharomyces cerevisiae can effectively increase the content of S7P to increase the yield of Gadusol. Therefore, we introduced the xylose utilization pathway into Saccharomyces cerevisiae and constructed a Saccharomyces cerevisiae strain that could use xylose as a carbon source.
Xylose, as a cheap carbon source, usually cannot be directly used by yeast. The xylose utilizing strain we constructed expanded the range of carbon sources available to Saccharomyces cerevisiae. Besides improving the yield of gadusol, it may reduce the cost of industrial production.
We can provide this Saccharomyces cerevisiae strain to more teams with similar needs. This strain can be used as chassis cells to produce other substances for iGEM teams in the future. You can also click here to get the gene sequence of all parts we used this year.

Fig.1a Metabolic Pathway Of Recombinant Saccharomyces cerevisiae

Fig.1b The Growth Curves Of Two Xylose Utilization Yeast
2. Sunscreen: Gadusol
We experimentally produced a substance called gadusol with UV absorption function and successfully extracted it from recombinant Saccharomyces cerevisiae by HPLC. Gadusol may meet the needs of some iGEM teams for UV protection in future experiments.

Fig.2a The Testing Result By HPLC
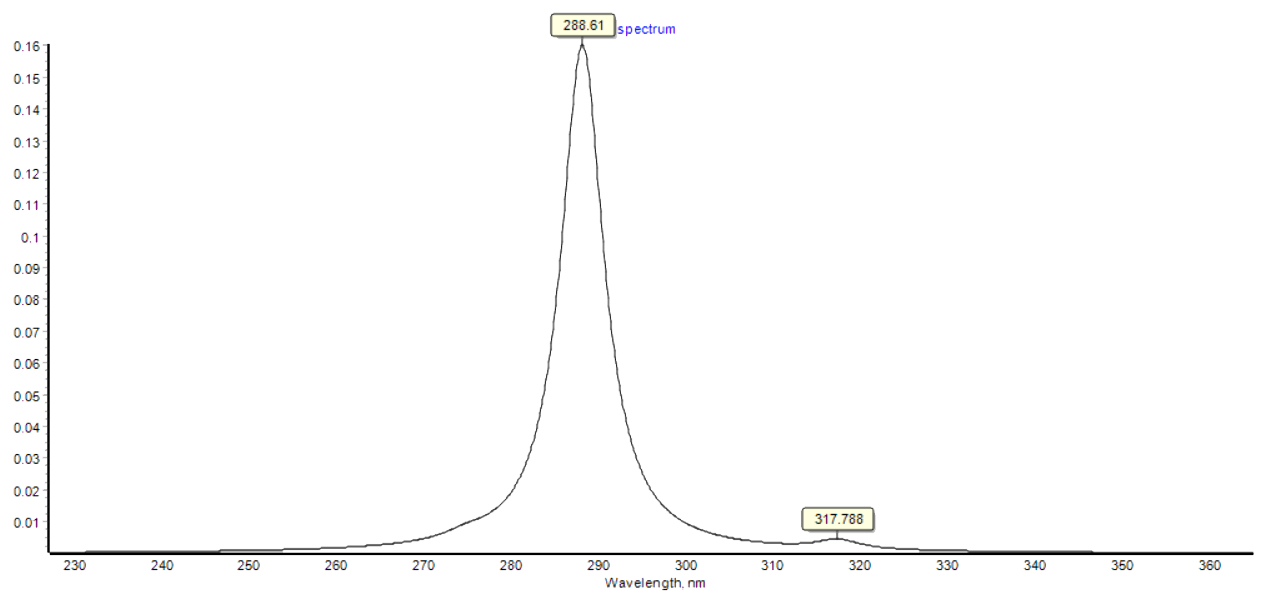
Fig.2b UV absorption spectrum stimulated by Chemdraw
3. Yeast Alliance
UESTC-China invited us to become a member of the Yeast Alliance. In the Alliance, we often share the molecular knowledge, fermentation, and practical experience related to Saccharomyces cerevisiae and solve experimental puzzles.
At present, we have finished an e-book based on our projects and experiments, and we decided to continue the "Yeast Alliance" in future iGEM competitions. We will actively update and share our experiences on the e-book for future iGEM teams using yeast as chassis organisms.

Fig.3 The Yeast Alliance Meetup: We Discuss & Exchange Our Experiences Freely
4. Information Update For Existing Part
In our project, we need to express three genes on one plasmid. In the process of consulting the literature, we found an article called "A Cas9 based toolkit to program gene expression in Saccharomyces cerevisiae", which described in detail the relative strength of different promoters in different media and culture time. In the follow-up experiments, we referred to this literature, selected the promoter of appropriate intensity, and achieved good results. So we share this article at the page BBa_K2365515, hoping to give a reference to our friends who haven't read this article yet.
This year, we used Metabolic Flux Analysis (MFA) Model to simulate metabolic flux changes and predict production after different molecular modifications, and then guide our wet lab. Besides, during the competition, we found several teams with similar needs failed to evaluate and predict due to the complexity of metabolic network. Thus, we would like to share the method we used to simplified the whole metabolic network and related equations to benefit future iGEM teams.
The number of iGEM teams in China is large and geographically dispersed. Small-scale meetup has a sizable geographical limit during the collaboration, which is not conducive to in-depth communication between teams. Therefore, in the early stage of the competition, we decided to hold a nationwide online meeting, cooperating with CCiC and three other teams located in different regions of China, and successfully hosted the 2021 China iGEM Online Meetup.
Compared to the national CCiC offline conference, usually held at the end of August, the meetup started earlier, making it easier for teams to exchange project designs, find partners and achieve long-term cooperation.

Fig.4 2021 China iGEM Online Meetup
Through communication with CCiC, we plan to continue holding such online meetups annually to provide communication opportunities for more teams.
As one of the main organizers, we made the plan, the manual, etc., and established the China iGEM Online Meetup WeChat group at this conference. These can be useful for future host teams, and the WeChat group can be a good platform for more teams to communicate.
To present the situation of coral bleaching and publicize synthetic biology knowledge, we will produce a "Glowing Glowing Gone" brochure and school textbooks and reach cooperation with Better Blue, Friends of Nature to expand influence.
Therefore, we can provide brochures and promotional platforms for future iGEM teams with projects related to coral conservation, environmental protection, and so on.




