<!DOCTYPE html>
Design
Cyanobacteria - the autotrophic module
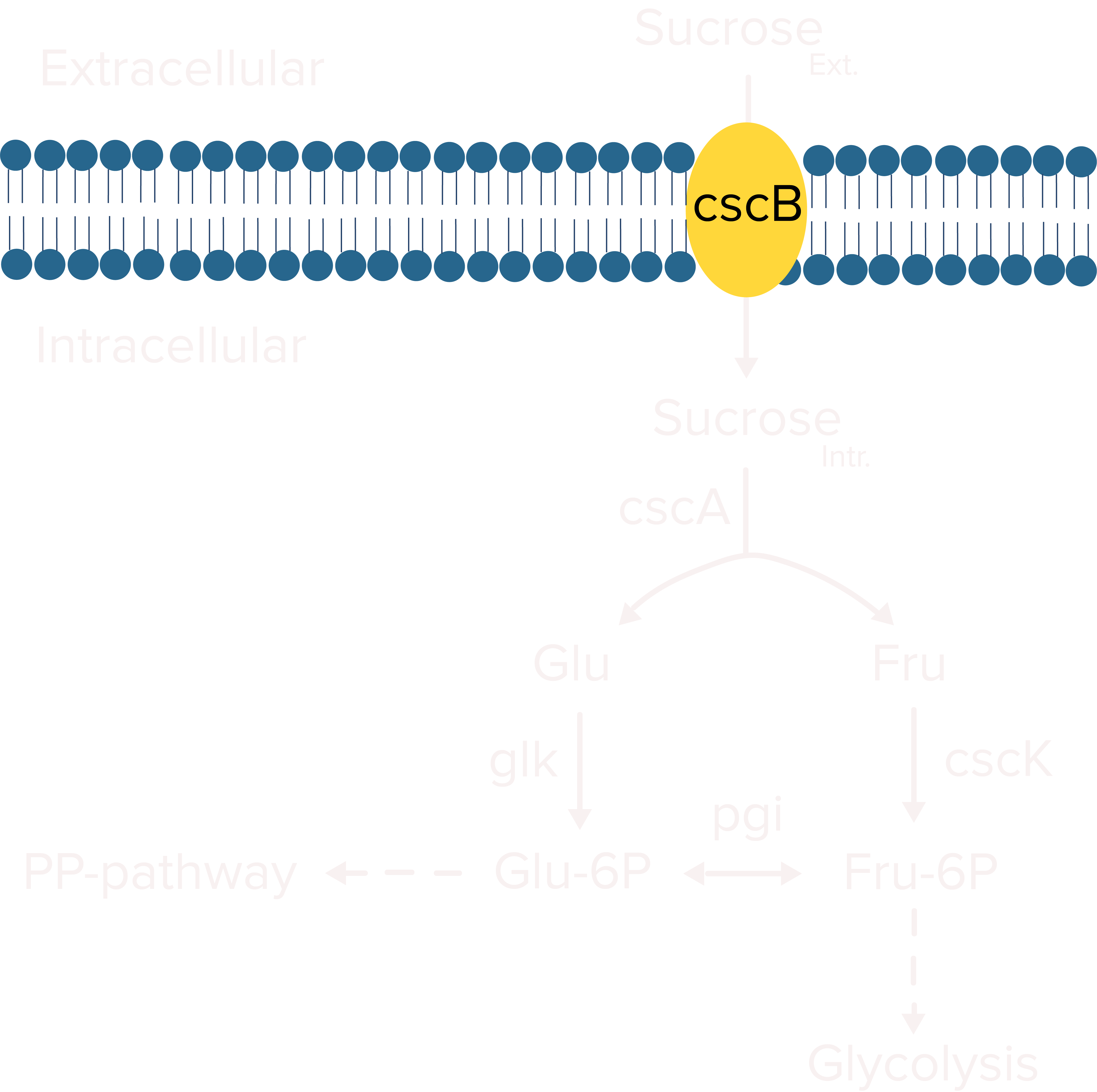
Certain cyanobacteria strains, like Synechococcus elongatus, naturally accumulate sucrose intracellularly under salt stress[1] to counter the osmotic pressure of their saline environment. We aimed to engineer S. elongatus UTEX 2973, a fast-growing strain of cyanobacteria,[2] to continually produce and secrete the sucrose it accumulates. To accomplish this, we intended to express cscB in our cyanobacterium. cscB, native to E. coli W, is a gene that codes for sucrose permease, a symporter that can transport sucrose and protons across the membrane of bacterial cells.[3]

The metabolic pathway for sucrose production in Synechococcus elongatus. Figure adapted from Lin et al. (2020)[7]
Abbreviations:
| ADP-Glu | adenosine diphosphate glucose |
| CBB cycle | Calvin-Benson-Bassham cycle |
| CscB | sucrose permease |
| F6P | fructose 6-phosphate |
| G1P | glucose 1-phosphate |
| G6P | glucose 6-phosphate |
| S6P | sucrose 6-phosphate |
| UDP-Glu | uridine diphosphate glucose |
| glgA | glycogen synthase |
| glgC | ADP-glucose pyrophosphorylase |
| sps | sucrose-phosphate synthase |
| spp | sucrose-phosphate phosphatase |
Once the symporter is present, it acts as a ‘sucrose sink’, continually transporting sucrose into the medium along the pH gradient, compelling the cyanobacterial cells to continually produce more sucrose to maintain their tonicity under salt stress.[3] This secreted sucrose would serve as a carbon source for the heterotrophic E. coli present in our co-culture setup.
We planned to culture the cyanobacteria in an alkaline, carbon-free, saline medium, to enable it to fix gaseous carbon dioxide as sucrose, and secrete it out. To maximize the yield of sucrose generated, we wished to express cscB under a range of constitutively expressed, strong promoters - including Pcpc560 (native to Synechocystis PCC 6803), J23119 (native to E. coli), PcpcB-m6 (a synthetic mutant variant of the native PcpcB) - and native promoters PpsbA2 and PpsbA3.[4][5][6]
Since S. elongatus UTEX 2973 is not naturally transformable, it is typically engineered using bacterial conjugation via triparental mating.[2] We opted for a CRISPR/Cas 12a (also known as CRISPR/Cpf1) based mechanism to integrate cscB into the Neutral Site 1 of UTEX 2973’s genome. The more commonly used Cas9 has been found to be toxic to cyanobacteria and hence cannot be used to modify it.[7]
Additionally, we also wished to look into the possible overexpression of the rate-limiting enzymes of the sucrose production pathway - sucrose phosphate synthase (SPS) and sucrose phosphate phosphatase (SPP). In most cyanobacteria these are coded for by the sps and spp genes respectively,[8] but a single sps gene codes for a fusion protein of the two in S. elongatus[16]. A similar CRISPR/Cas 12a based approach would allow for the genomic integration of sps into the Neutral Site 3 of UTEX 2973’s genome.
However, we were pressed for time due to limited lab access during the ongoing pandemic and were unable to carry out all the experiments we designed. For a proof of concept, we used the pre-engineered strains of S. elongatus UTEX 2973 kindly given to us by Prof. Pakrasi, at the Washington University at St. Louis.[7]
We received the following strains from them:
- S. elongatus UTEX 2973 WT
- S. elongatus UTEX 2973-cscB
- S. elongatus UTEX 2973-cscB-sps
- S. elongatus UTEX 2973-cscB-sps-spp
Due to limited time and the constraint of just a single incubator, we worked primarily with the first and second strains. The WT was chosen as a negative control for all our experiments.The reasoning behind our choice of sucrose-producing strain was the observation by Lin et al., 2020[7], that the cscB strain had the highest yields of sucrose. We do however plan on working with the other strains in future.
In UTEX 2973-cscB, cscB was integrated into the Neutral Site 3 of the cyanobacterium’s genome under a lacUV5 promoter, using a suicide vector. In the latter two strains, sps and spp genes from the cyanobacterium Synechocystis PCC 6803 were respectively expressed under a trc10 promoter, through a self-replicating RSF1010 based plasmid. They have achieved a maximum sucrose titer of 8.1 g/L over 5 days and a maximum daily productivity rate of 1.9 g/L/day in the 2973-cscB strain.[7]
E. coli - the heterotrophic module
E. coli is commonly used for industrial chemical biosynthesis. However, most commonly used strains of E. coli cannot naturally consume sucrose.[9] This module focuses on engineering E. coli to take up sucrose produced by S. elongatus in the co-culture and metabolize it to produce industrially important metabolites.
Our initial goal was to produce succinic acid as a final product through E. coli under aerobic conditions, by engineering its metabolic pathways. The aerobic conditions would be facilitated by oxygen evolved by S. elongatus. However, we decided against succinate production as we would not have enough time to engineer E. coli’s metabolic pathways from scratch, and also due to the inputs we received about the advantages of microaerobic conditions over aerobic and anaerobic conditions for metabolite production, from Dr. Yazdani
As a proof of concept, we chose instead to work with E. coli KJK01, provided to us by Dr. Yazdani. This is a modified MG1655-based strain that can metabolize glucose and xylose to produce butanol.
E. coli KJK01 produces optimal butanol yields under microaerobic conditions, and also produces equal quantities of ethanol and some amount of pyruvate. This strain would have to be modified to consume sucrose so that it may be cultivated alongside S. elongatus in our co-culture design.
Sucrose Transport and Metabolism:
E. coli W is one of few non-pathogenic wild-type strains that can naturally consume sucrose. It does so with the help of three crucial genes:11
- cscB: codes for sucrose permease, a symporter that transports sucrose and protons across the bacterial cell membrane.
- cscA: codes for sucrose-6-phosphate hydrolase (invertase) that hydrolyses sucrose into glucose and fructose.
- cscK: codes for fructokinase that catalyzes the conversion of fructose to fructose-6-phosphate.
that the WT strain, lacking cscB can act as a negative control for all our experiments
Sucrose consumption pathway in E. coli W. Figure adapted from Mohamed et al., (2019)[11]
Claudia Vickers et al. (2012) created pCSCX, a plasmid containing these three genes under a native bidirectional promoter.12 When transformed into KJK01, the plasmid would enable the bacterium to subsist on sucrose as its only available source of carbon. The transformed strain, pCSCX-KJK01 will be ready for a co-culture with S. elongatus, consuming the sucrose generated by it to produce butanol.

Map of the pCSCX plasmid adapted from Bruschi et al. (2012)[12]
Butanol production
The KJK01 strain uses the acetoacetyl-CoA reduction pathway to produce one molecule of butanol from two molecules of acetyl-CoA.10 The strain carries:
- From Clostridium acetobutylicum:
- hbd - codes for beta-hydroxybutyryl-CoA dehydrogenase which converts acetoacetyl-coA to (S)-3-hydroxybutanoyl-CoA.
- crt - codes for 3-hydroxy-butyryl-CoA dehydratase that converts (S)-3-hydroxybutanoyl-CoA to crotonyl-CoA.
- adhE2 - codes for butyraldehyde/butanol dehydrogenase, which is a multifunctional enzyme with both alcohol dehydrogenase and acetaldehyde dehydrogenase activities.
- From Treponema denticola
- ter - codes for trans-enoyl-coenzyme A (CoA) reductase that converts acyl-CoA to trans-2,3-dehydroacyl-CoA
- From E.coli
- atoB - coding for acetyl-CoA acetyltransferase that converts acetyl-coA to acetoacetyl-coA.
These genes were integrated using CRISPR/Cas9 into the genome in a markerless fashion.[10]
Additionally, it has the following knockouts:
- pta - codes for an enzyme that catalyzes the conversion of acetyl-CoA to acetate.
- adhE - codes for an enzyme that catalyzes the conversion of acetyl-CoA to ethanol.
- frdA - codes for an enzyme that catalyzes the conversion of fumarate to succinate.
- ldhA - codes for an enzyme that catalyzes the conversion of pyruvate to lactate.

Butanol production pathway in E. coli KJK01 with associated knockouts (represented by an X).Figure adapted from Abdelaal et al., 2019[10]
The above pathway is a Clostridial-based fermentative pathway for n-butanol production. Three of the genes are from Clostridium, with the exception of the first enzyme THL from E. coli itself and the intermediate enzyme TER from the T. denticola. These alternative enzymes from non-Clostridial sources were shown to be more efficient in n-butanol production via fermentation.10
In order to make this strain capable of consuming sucrose from the medium, we will express cscB gene, coding for the sucrose transporter, in it. This sucrose then will be converted to glucose and fructose via cscA and cscK, of which the former enters the butanol synthesis pathway in the engineered E. coli strain.
E. coli KJK01 strain generated a yield of 5.4g/L butanol over 27 hours from glucose and we hope to get a similar yield with sucrose.[10] After sufficient assays confirming that pCSCX-KJK01 is capable of surviving with sucrose as its sole carbon source, it will be co-cultured with engineered S. elongatus to validate the co-culture.
You can read more about the constructs we made and the experiments we performed with these strains here and here.
Succinate production
Our original plan was to modify an E. coli straincapable of sucrose metabolism to produce succinate via the reductive branch of the TCA cycle, activated during anaerobic respiration. One of the main limiting factors of this process is the availability of NADH, with one mole of succinate requiring two moles of NADH.13 Thus four knockouts are made to get rid of pathways competing for NADH. An additional knockout is made to activate the glyoxylate pathway, so that succinate is now produced through two routes (see Fig 4). A non-native pyruvate carboxylase coding gene pyc is expressed to reduce pyruvate accumulation in the cell.[13]
These modifications have previously been shown to produce a yield of 1.59 moles of succinate per mole glucose, 13 and 3.33 moles of succinate per mole sucrose.[14]
The list of modifications is as follows:
- Knockout ldhA
- Knockout adhE
- Knockout ack-pta
- Knockout iclR
- Overexpress non-native pyc
The final pathway after these modifications is as follows:

Modified metabolic pathway in E. coli which produces succinate as its final product. X on a gene name represents a knockout of that gene.
Modelling
Our modeling objective was primarily to optimize butanol production. This involved predicting individual growth rates and the production yields of our organisms, identifying key gene modification targets, and analyzing crucial co-culture parameters.
To begin modeling, we needed to make adjustments to the available models of E. coli and S. elongatus 2973 - iML1515 and iSyu683 respectively- , to match the strains that we would be using in our co-culture. This involved adding reactions and knockouts that weren’t already present in the base models. For S. elongatus, we additionally had to model salt stress, the condition responsible for sucrose production in the first place.
To simulate the growth rates, sucrose, and butanol production in monocultures, we planned to use two constraint-based analysis techniques - Flux Balance Analysis (FBA) and Minimum of Metabolic Adjustments (MoMA). Knowing how the individual strains react to various environmental conditions would also serve to validate the results of our co-culture modeling.
We then planned to analyze the co-culture to identify optimal parameter values for maximum butanol production. This would be done using two techniques - SteadyCom and dynamic modeling, which we performed in collaboration with Team Toulouse_INSA_UPS.
SteadyCom is used to analyze the steady-state behavior of co-cultures, microbiomes, and host-microbe interactions15. It provides results such as the biomass ratio of the constituent organisms at steady-state growth, and the fluxes of various reactions in the multi-species network. It would allow us to identify certain key metabolites that play an important role in the interaction between the organisms.
Dynamic modeling shows the time evolution of concentrations of metabolites and abundances of the organisms in the co-culture in the bioreactor setup.
We then identify key gene modifications in the individual organisms E. coli and S. elongatus that would allow us to achieve these optimal values. This would be done using OptKnock and Flux Scanning based on Enforced Objective Flux (FSEOF).
FSEOF is used to identify metabolic reactions involved in sucrose and butanol production and correspondingly find the genes that regulate them.[17] These genes can be overexpressed to increase sucrose production in S. elongatus and butanol production in E. coli.
OptKnock on the other hand is an algorithm that finds and suggests which set of reactions to remove from a metabolic network to obtain a mutant that will produce a particular target of interest at a higher rate than the wild-type strain.[18]
The final results obtained from all the analyses would provide us with crucial bioreactor parameters and gene modifications in the individual organisms E. coli and S. elongatus that would allow us to achieve optimal butanol production, and inform us of productive avenues to explore in the lab.
References
- Klähn, S., & Hagemann, M. (2011). Compatible solute biosynthesis in cyanobacteria. Environmental microbiology, 13(3), 551-562.
- Yu, J., Liberton, M., Cliften, P. et al. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci Rep 5, 8132 (2015). https://doi.org/10.1038/srep08132
- Ducat, D. C., Avelar-Rivas, J. A., Way, J. C., & Silver, P. A. (2012). Rerouting carbon flux to enhance photosynthetic productivity. Applied and environmental microbiology, 78(8), 2660–2668. https://doi.org/10.1128/AEM.07901-11
- Vasudevan R, Gale GAR, Schiavon AA, et al. CyanoGate: A Modular Cloning Suite for Engineering Cyanobacteria Based on the Plant MoClo Syntax. Plant Physiol. 2019;180(1):39-55. https://doi.org/10.1104/pp.18.01401
- Sengupta, Annesha, Swati Madhu, and Pramod P. Wangikar. "A Library of tunable, portable, and inducer-free promoters derived from cyanobacteria." ACS Synthetic Biology 9.7 (2020): 1790-1801. https://doi.org/10.1021/acssynbio.0c00152
- Li S, Sun T, Xu C, Chen L, Zhang W. Development and optimization of genetic toolboxes for a fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Metab Eng. 2018 Jul;48:163-174. https://doi.org/10.1016/j.ymben.2018.06.002
- Lin, PC., Zhang, F. & Pakrasi, H.B. Enhanced production of sucrose in the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Sci Rep 10, 390 (2020). https://doi.org/10.1038/s41598-019-57319-5
- Ungerer, J., Pakrasi, H. Cpf1 Is A Versatile Tool for CRISPR Genome Editing Across Diverse Species of Cyanobacteria. Sci Rep 6, 39681 (2016). https://doi.org/10.1038/srep39681
- Sabri, S., Nielsen, L. K., & Vickers, C. E. (2013). Molecular control of sucrose utilization in Escherichia coli W, an efficient sucrose-utilizing strain. Applied and environmental microbiology, 79(2), 478–487. https://doi.org/10.1128/AEM.02544-12
- Abdelaal, A. S., Jawed, K., & Yazdani, S. S. (2019). CRISPR/Cas9-mediated engineering of Escherichia coli for n-butanol production from xylose in defined medium. Journal of Industrial Microbiology and Biotechnology, 46(7), 965-975. https://doi.org/10.1007/s10295-019-02180-8
- Mohamed, E.T., Mundhada, H., Landberg, J. et al. Generation of an E. coli platform strain for improved sucrose utilization using adaptive laboratory evolution. Microb Cell Fact 18, 116 (2019). https://doi.org/10.1186/s12934-019-1165-2
- Bruschi, M., Boyes, S. J., Sugiarto, H., Nielsen, L. K., & Vickers, C. E. (2012). A transferable sucrose utilization approach for non-sucrose-utilizing Escherichia coli strains. Biotechnology advances, 30(5), 1001–1010. https://doi.org/10.1016/j.biotechadv.2011.08.019
- Ailen M. Sánchez, George N. Bennett, Ka-Yiu San, Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity, Metabolic Engineering, Volume 7, Issue 3, 2005, Pages 229-239, ISSN 1096-7176, https://doi.org/10.1016/j.ymben.2005.03.001
- Wang J, Zhu J, Bennett GN, San KY. Succinate production from different carbon sources under anaerobic conditions by metabolic engineered Escherichia coli strains. Metab Eng. 2011 May;13(3):328-35. Epub 2011 Mar 30. PMID: 21440082. https://doi.org/10.1016/j.ymben.2011.03.004
- Chan, S. H. J., Simons, M. N., & Maranas, C. D. (2017). SteadyCom: predicting microbial abundances while ensuring community stability. PLoS computational biology, 13(5), e1005539.
- Duan, Y., Luo, Q., Liang, F. et al. Sucrose secreted by the engineered cyanobacterium and its fermentability. J. Ocean Univ. China 15, 890–896 (2016).https://doi.org/10.1007/s11802-016-3007-8
- Choi, H. S., Lee, S. Y., Kim, T. Y., & Woo, H. M. (2010). In silico identification of gene amplification targets for improvement of lycopene production. Applied and environmental microbiology, 76(10), 3097–3105. https://doi.org/10.1128/AEM.00115-10
- Burgard, A. P., Pharkya, P., & Maranas, C. D. (2003). Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnology and bioengineering, 84(6), 647–657. https://doi.org/10.1002/bit.10803









Team IISER Pune India