<!DOCTYPE html>
Contribution
Parts data
Among the many contributions we made this year, we want to emphasize the new experimental data we obtained to characterize the pLux promoter (Part BBa_K2656003 which is the Golden Braid compatible, together with its Biobrick standard sister part BBa_R0062) in the context of the transcriptional unit built in part BBa_K2656122.
Experimental data from pLux promoter
Following a standard experimental procedure and the IGEM 2018 Interlab calibration for Particles and MEFL we obtained the temporal behavior of the device for different inducer concentrations.

Final GFP expression levels for different AHL inducer concentrations

We used this data (click here to get the raw data) to characterize the Golden Braid version (BBa_K2656003) of pLux promoter (BBa_R0062).
Characterization of pLux promoter
Using the same algorithm and methods as Valencia_UPV team, we have obtained the parameters for the pLux promoter in the context of the following model and experimental conditions:
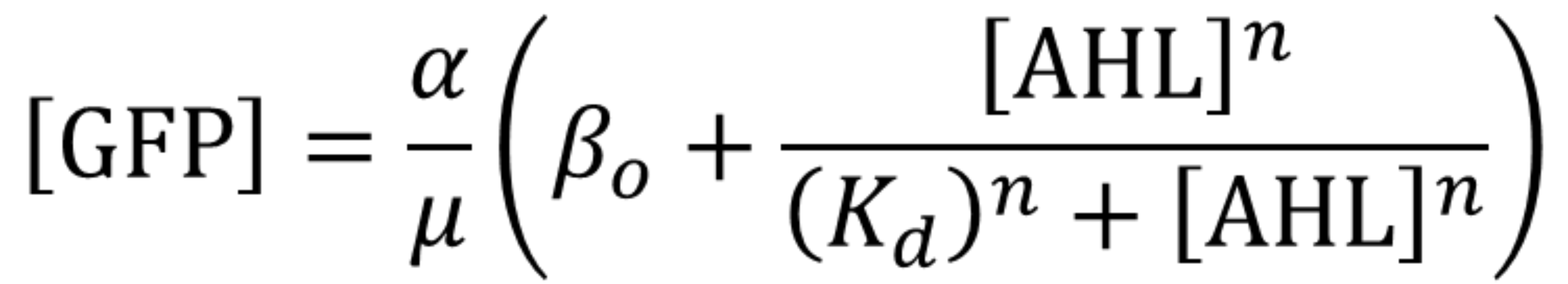
| Parameter | Value |
| Effective translation rate 𝛼 | 𝛼 = 8.11e8 molec min-1 |
| Effective dissociation constant Kd | Kd = 15.34 nM |
| Hill coefficient | n = 0.958 |
| Basal expression | 𝛽 = 0.1049 |
| Growth (Dilution) rate μ | μ = 0.0115 min-1 |
Using this optimized parameter values we obtained the simulated hill function corresponding to the previous equation and compared it with the experimental data obtained (for steady-state, this is after reaching equilibrium).
The characterization of this part, and in particular the parameters that are from the promoter itself: Effective dissociation constant Kd, Hill coefficient, and Basal expression was used in our DBTL cycle for development of Module 2, and also this information played a tremendous role in the success and prediction power of our modeling.

Materials and Methods
- All experimental measurements were taken with Biotek CytationTM 3, using a 96-well plate.
- The cells were E. cloni® cells (Lucigen) transformed with the corresponding plasmid.
- For all experiments, we worked with an Glycerol stocks that contains the corresponding plasmid.
- Cultures of 3mL falcon tubes with sLB medium with the corresponding antibiotic were prepared and incubated overnight (18 hours) at 37°C, 200 rpm.
- The overnight cultures were refreshed in 3 mL of Minimal Media M9 with the corresponding antibiotic and incubated at 37°C, 200 rpm for 4 hours.
- Culture tubes at OD600=0.1 were sit in cold water for 30 mins.
- We measured absorbance and fluorescence of four replicas of the sample starting with OD600=0.1 during 1 hour.
- We induced with the approapriate concentration of AHL to get desired final conentrations, and continued measuring absorbance and fluorescence during 8 hours.
- PROTOCOL: Time between measures 5 min, Temperature 37°C, Shaking Double orbital (Continuously), Absorbance wavelength 600 nm, Excitation wavelength 485 nm, Emission wavelength 528 nm, 96-well volume (individual) 200 μl.
Note: This characterization data for part R0062 (BBa_K2656003) needs to be interpreted in the context of the transcriptional units from where the experimental data was taken.
DYO DEVICES
Within the Igem Ecuador team, two devices were made, a fluorometer and a transilluminator, which were built with fast machining techniques such as laser cutting and 3D printing. The use of these techniques allowed us to lower the manufacturing costs of the devices at an affordable cost and with easy-to-find materials. Another important aspect was the implementation of IOT methodologies which allowed us to control and monitor the equipment from a phone through a wifi network. These technologies will allow us to lay the foundations for the new devices to allow us to have facilities such as remote control or remote viewing from remote areas. For details of the devices, you can find them in the following link
Modular platform for dsRNA production
Agrobactory 593 was designed as a modular platform to produces double-stranded RNA (dsRNA) of one or several genes from the pathogen of interest genome. Once inside the pathogen, this dsRNA is further processed into RNA interference that can regulate the expression of any gene in the genome. Therefore, these RNA molecules can become a biopesticide to treat plant diseases. Our collection has two groups of parts, the specific dsRNA to prevent Fusarium wilt by Foc TR4 (as an example of what can be done) and the parts that are necessary to build the modular platform. This second groups is a library of constructs with the T7 promoter and combinations of RBSs and Shine-Delgarno sequences. Once assembled, they become a destination vector (promoter, rbs, recognition sites and terminator in both forward and reverse directions) that is able to receive dsRNA sequences in the desired location when digested with the appropriate enzymes. For details of the design, you can find them in the following link
Synthetic Biology Club, support to new teams Ec
In order to inspire students in science and Synthetic Biology, we created a club within our institution to show interest in those who did not know about this area, and support those who have already begun to explore Synthetic Biology. The purpose is to lay the foundations of this area so little known in our country, to continue forming teams within our university and other institutions in Ecuador. Detailed information about the Synthetic Biology Club and support for the formation of new teams can be found at Synthetic Biology at the University


